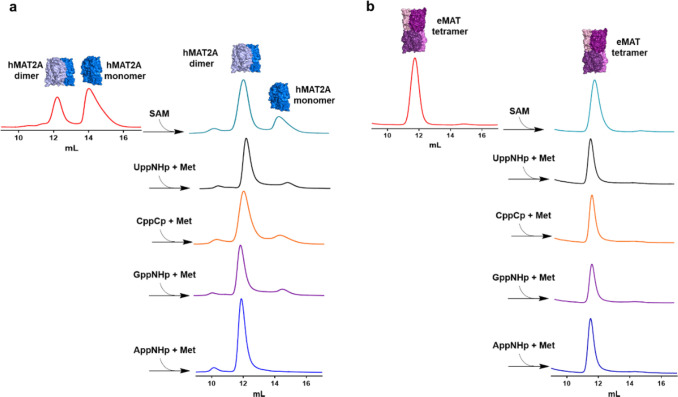
Figure 2.
Analysis of oligomeric state of hMAT2A and eMAT by size-exclusion chromatography. (a) hMAT2A (20 μM) is incubated with non-hydrolyzable NTPs (1 mM) adenosine-5′-[(β, γ)-imido]triphosphate (AppNHp), guanosine-5′-[(β, γ)-imido]triphosphate (GppNHp), cytidine-5′-[(β, γ)-methyleno] triphosphate (CppCp), and uridine-5′-[(β, γ)-imido]triphosphate (UppNHp)] together with methionine (Met, 10 mM) using reaction buffer (100 mM HEPES, 10 mM KCl, 10 mM MgCl2, pH 8, 37 °C for 1 h). hMAT2A is in an equilibrium of a monomer and dimer. When incubated with both substrates, the enzyme converts completely to a dimeric state. No change in oligomeric state is observed when incubated with SAM alone. (b) eMAT (20 μM) is incubated using the same conditions as used for hMAT2A. eMAT is in a tetrameric state, and no change in oligomeric state was observed after incubation with both substrates and SAM. Size-exclusion chromatography was performed using a GE Healthcare Life Sciences using Superdex 200 Increase 10/300 GL column.