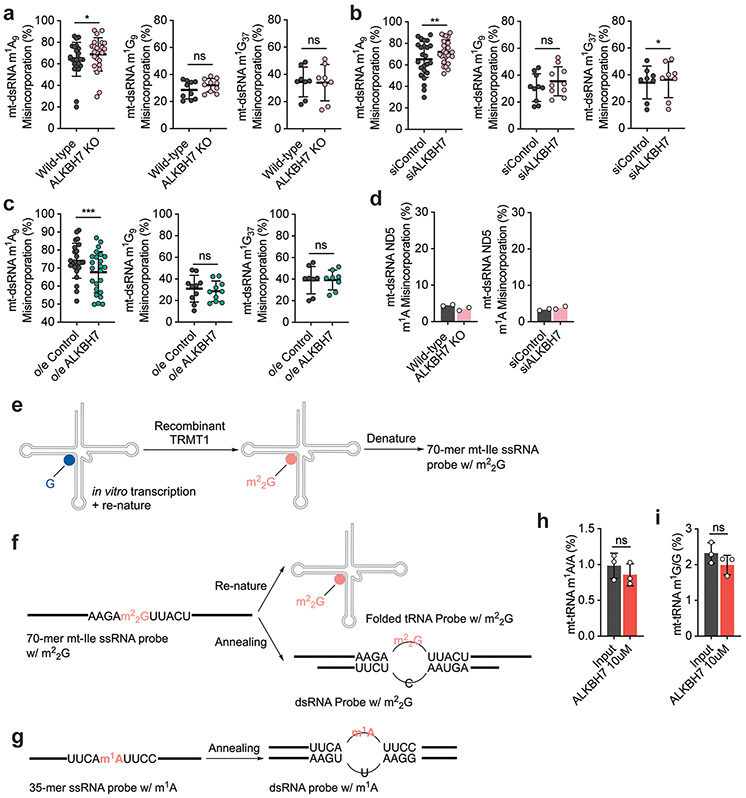
Extended Data Fig. 4 ∣. ALKBH7 demethylation effect on other methylations (such as m1A9, m1G9, and m1G37) within mitochondrial dsRNA is lower than that on Ile-m22G and Leu1-m1A inside cells.
a, Mutation levels for m1A9, m1G9, and m1G37 in mt-dsRNA in ALKBH7 knockout HepG2 cells vs. wild-type. m1A9: *P = 0.0398; paired, two-tailed t-test; n = 24, 12 independent m1A sites. m1G9: P = 0.0954; paired, two-tailed t-test; n = 10, 5 independent m1G sites. m1G37: P = 0.38; paired, two-tailed t-test; n = 8, 4 independent m1G sites. b, Mutation levels for m1A9, m1G9, and m1G37 in mt-dsRNA in siALKBH7 HepG2 cells vs. siControl. m1A9: **P = 0.0047; paired, two-tailed t-test; n = 24, 12 independent m1A sites. m1G9: P = 0.1462; paired, two-tailed t-test; n = 10, 5 independent m1G sites. m1G37: *P = 0.0188; paired, two-tailed t-test; n = 8, 4 independent m1G sites. c, Mutation levels for m1A9, m1G9, and m1G37 in mt-dsRNA in ALKBH7-overexpressed HeLa cells vs. control. m1A9: ***P = 0.0002; paired, two-tailed t-test; n = 24, 12 independent m1A sites. m1G9: P = 0.5666; paired, two-tailed t-test; n = 10, 5 independent m1G sites. m1G37: P = 0.4838; paired, two-tailed t-test; n = 8, 4 independent m1G sites. For a-c, each data point represents one methylation site, with the misincorporation from 2 biologically independent samples. Data are presented as mean values ± SD. d, Mutation levels for m1A in mt-ND5 region of mt-dsRNA. Left: ALKBH7 knockout HepG2 cells vs. wild-type; Right: siALKBH7 HepG2 cells vs. siControl. n = 2 biologically independent samples. e, m22G-containing ssRNA probe synthesized by recombinant TRMT1 protein. f, m22G-containing mt-Ile RNA probe in forms of folded tRNA and dsRNA. g, m1A-containing mt-Leu1 RNA probe in forms of ssRNA and dsRNA. h, Modification levels of m1A/A in purified mt-tRNA before and after treatment with recombinant human ALKBH7. P = 0.4048; unpaired, two-tailed t-test. i, Modification levels of m1G/G in purified mt-tRNA before and after treatment with recombinant human ALKBH7. P = 0.2256; unpaired, two-tailed t-test. For h-i, n =3 biologically independent samples. Data are presented as mean values ± SD.