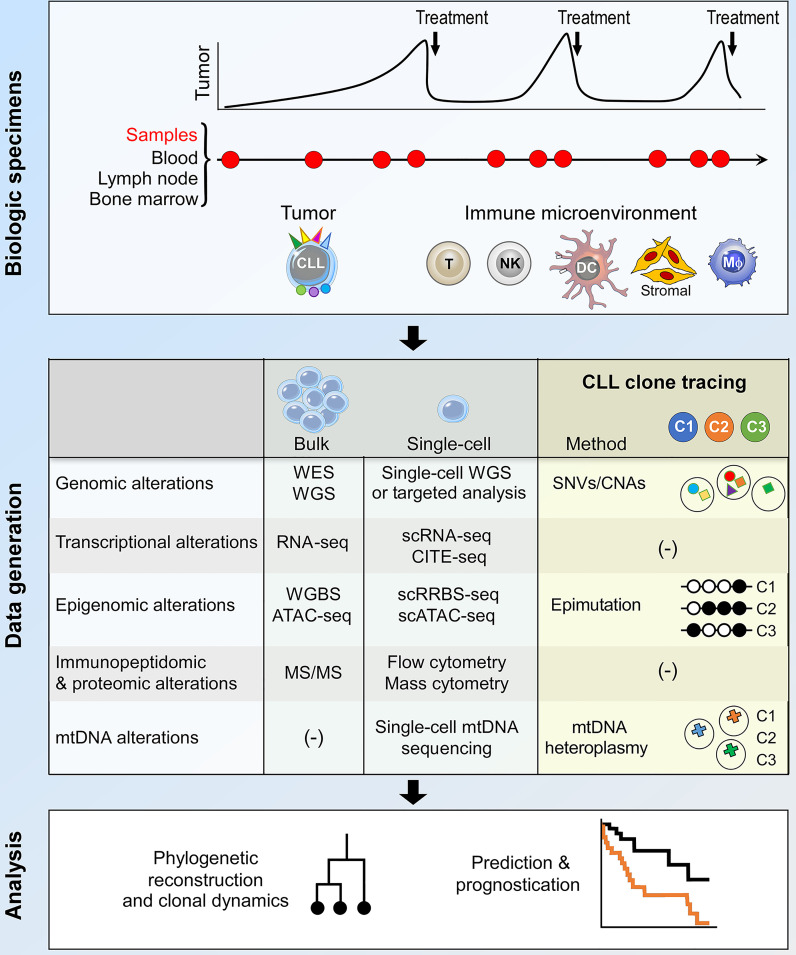
Figure 2.
Integrative bulk and single-cell analysis of CLL clonal dynamics. The current approach to the study of CLL clonal evolution is summarized, highlighting the use of biological specimens (top panel) and methodologies for data generation and analysis (middle and bottom panels respectively). Longitudinal samples collected at various time points during the clinical course of a patient allow analysis of both the tumor and the immune microenvironment. Various bulk and single-cell data generation approaches can be used to interrogate biological alterations that underpin clonal evolution. Within the context of single-cell analysis, lineage tracing techniques facilitate the identification of CLL subclones and the integration of multimodal data pertaining to individual subclones. The data thus generated can be used for phylogenetic reconstruction, analysis of subclonal dynamics and clinical prognostication. DC, dendritic cell; Mφ, macrophage; WES, whole exome sequencing; WGS, whole genome sequencing; SNV, single-nucleotide variation; CNA, copy number alteration; mtDNA, mitochondrial DNA; scRNA-seq, single-cell RNA-seq; CITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; WGBS, whole genome bisulfite sequencing; scRRBS-seq, single-cell reduced representation bisulfite sequencing; scATAC-seq, single-cell sequencing assay for transposase-accessible chromatin; MS/MS, tandem mass spectrometry.