Fig. 2.
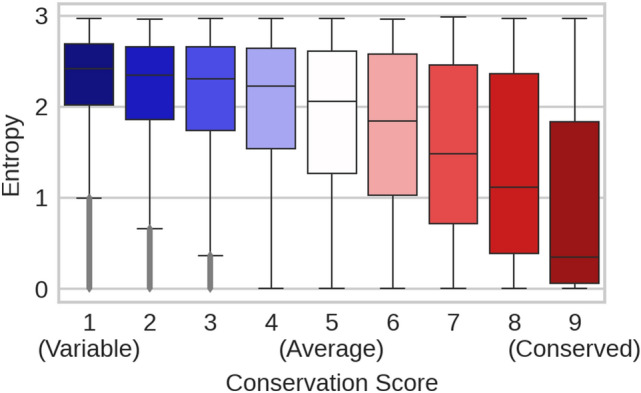
pLMs captured conservation without supervised training or MSAs. ProtBert was optimized to reconstruct corrupted input tokens from non-corrupted sequence context (masked language modeling). Here, we corrupted and reconstructed all proteins in the ConSurf10k dataset, one residue at a time. For each residue position, ProtBert returned the probability for observing each of the 20 amino acids at that position. The higher one probability (and the lower the corresponding entropy), the more certain the pLM predicts the corresponding amino acid at this position from non-corrupted sequence context. Within the displayed boxplots, medians are depicted as black horizontal bars; whiskers are drawn at the 1.5 interquartile range. The x-axis gives categorical conservation scores (1: highly variable, 9: highly conserved) computed by ConSurf-DB (Ben Chorin et al. 2020) from multiple sequence alignments (MSAs); the y-axis gives the probability entropy (Eq. 13) computed without MSAs. The two were inversely proportional with a Spearman’s correlation of -0.374 (Eq. 11), i.e., the more certain ProtBert’s prediction, the lower the entropy and the higher the conservation for a certain residue position. Apparently, ProtBert had extracted information correlated with residue conservation during pre-training without having ever seen MSAs or any labeled data
