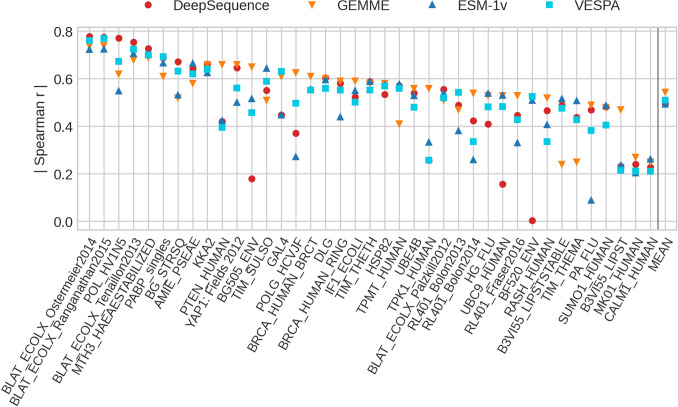
Fig. 5.
No SAV effect prediction consistently best on DMS data. Data: DMS39 (39 DMS experiments gathered for the development of DeepSequence (Riesselman et al. 2018)); experiments sorted by the maximum absolute Spearman coefficient for each experiment. Methods: a DeepSequence trained an unsupervised model for each DMS experiment using only MSA input, i.e., no effect score labels were used (Riesselman et al. 2018); b GEMME inferred evolutionary trees and conserved sites from MSAs to predict effects (Laine et al. 2019); c ESM-1v correlated log-odds of substitution probabilities (Methods) with SAV effect magnitudes (Meier et al. 2021); d VESPA (this work) trained a logistic regression ensemble on binary SAV classification (effect/neutral) using predicted conservation (ProtT5cons), BLOSUM62 (Henikoff and Henikoff 1992), and log-odds of substitution probabilities from ProtT5 (Elnaggar et al. 2021) as input (without any optimization on DMS data). The values for the absolute Spearman correlation (Eq. 11) are shown for each method and experiment. The rightmost column shows the mean absolute Spearman correlation for each method. Although some experiments correlated much better (toward left) with predictions than others (toward right), the spread between prediction methods appeared high for both extremes; DeepSequence was the only method reaching a correlation of 0 for one experiment; another one and three experiments were predicted with correlations below 0.2 for ESM-1v and DeepSequence, respectively, while the vast number of the 4 × 39 predictions reached correlations above 0.4