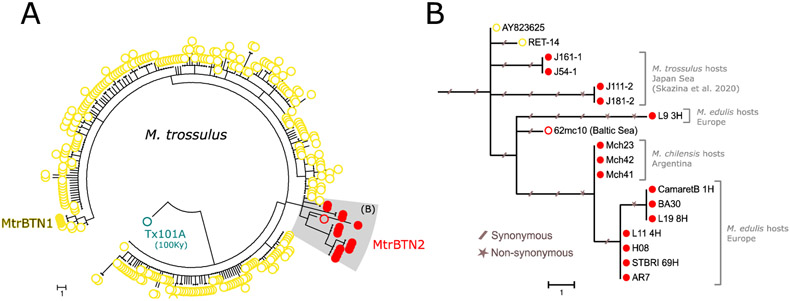
Fig 6: Phylogenetic analysis of mtCOI sequences.
(A) Maximum parsominy tree on mtCOI 603bp aligned sequences of our 5 MtrBTN2 tumor samples (red filled circles, L9_3, CamaretB_1, L19_8, L11_4, STBRI69) and all available M. trossulus (GenBank) including three MtrBTN1 tumor samples (yellow filled circles), seven MtrBTN2 tumor samples of which four are from M. trossulus hosts (Skazina et al. 2020) and three are from M. chilensis hosts (red filled circles), a 100 000 years old (Tx101A, blue open circles) and contemporary M. trossulus mussels. (B) Subtree of MtrBTN2 mtCOI haplotypes with the two closest relatives from healthy mussels. Synonymous and non-synonymous mutations are reported with slashed traits and stars respectively. Host species and geographical origins are indicated on the right side of the tree.