Figure 3.
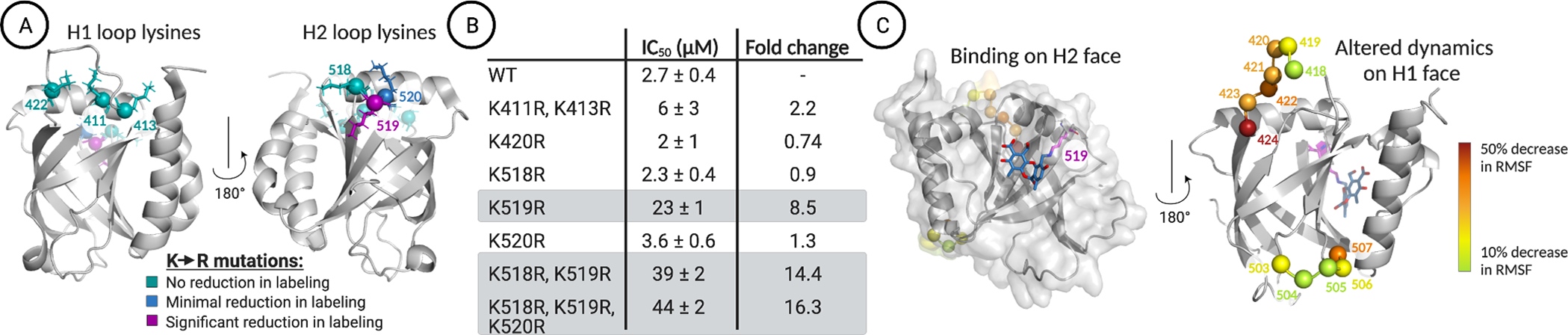
(A) LC–MS analysis of norstictic acid covalent adduct formation with Med25 lysine-to-arginine mutants indicates that K519R leads to the most significant reduction of labeling. No reduction of labeling corresponds to a decrease in abundance of the NA covalent adduct of less than 10%. Minimal reduction in labeling, observed for K520R, corresponds to a 22% reduction in the mass abundance of the NA covalent adduct. Significant reduction in labeling, observed for K519R, corresponds to a 53% reduction in the mass abundance of the +1 covalent adduct. The Supporting Information provides additional quantitative analysis. PDB entry 2XNF was used to generate the figure. (B) Inhibition of the Med25 AcID ETV5 interaction by NA measured using fluorescence polarization. Mutants containing K519R, highlighted in gray, demonstrate the most significant increase in apparent IC50. Values represent averages of three independent experiments with the indicated error (SDOM) (C) (left) Centroid structure of the most populated cluster from molecular dynamics simulations, where NA binds to the H2 face of Med25 and covalently links to K519. (right) The residues that showed the greatest reduction in fluctuations (RMSF) upon activator binding all occur on dynamic substructures on the H1 face.
