Figure 1.
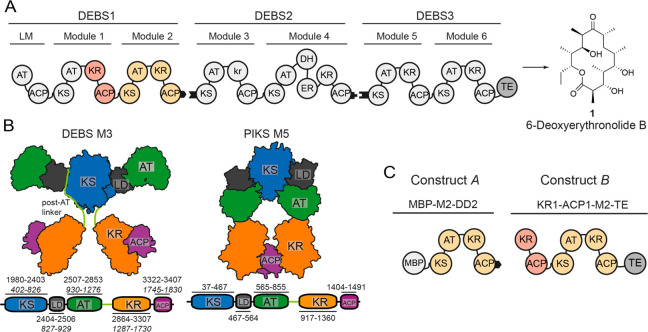
Schematic architecture of DEBS, structural models, and constructs used in this study. (A) Overview of DEBS. The three polypeptides (DEBS1–3), the encoded modules (M1–M6), the loading module (LM), the TE, and the final product are shown. Black tabs depict docking domains. The KR domain of M3 is redox-inactive and termed kr.33 (B) Models for the partially reducing PKS modules; DEBS M3 is based on SAXS analysis that implemented high-resolution structural information on KS-AT models and FAS,9 and PIKS M5 is based on cryoEM analysis.14 DEBS M3 model: Domains KS and AT form a rigid dimeric unit of an overall extended shape, and are separated by a noncatalytic linker-domain (LD). AT is followed by a structured post-AT linker10,11,34 that interacts with both LD and KS, and connects AT to KR. PIK M5 model: In the arch-shaped KS-AT dimer, the AT is oriented downward. It interacts with the respective upward-facing LD and the KS of the other protomer of the dimer. The post-AT linker is not resolved within the PIKS M5 structure. Schemes of the domain structures including domain boundaries are attached. Note that for DEBS M3, domain borders are also given in construct A numbering with labeling in italics, which has been used in this study (Table S1). Post-AT linker sequence as defined in refs (21 and 22). (C) DEBS M2-derived constructs MBP-M2-DD2 (construct A) and KR1-ACP1-M2-TE (construct B).