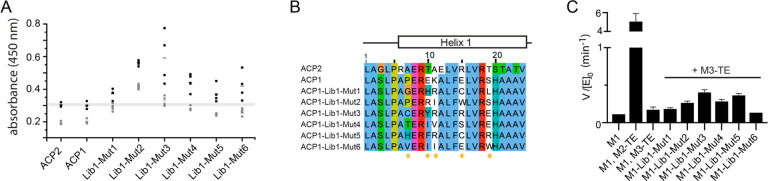
Figure 4.
Directed evolution of a chimeric ACP:KS interface. Analysis of enriched mutants from ACP1-Library1. (A) ELISA of ACPs presented on the phage surface. Results for selected mutants (Lib1-MutX) and wild-type ACPs are shown. Signal was obtained in KS3-AT3-coated wells (black bars) and the degree of unspecific binding was assessed by comparing it to the signal in BSA coated wells (gray bars). For each ACP, four individually grown phage cultures were tested (except three cultures for ACP1 and ACP2), and the individual data points and data mean are indicated. Data range of ACP2 is indicated by a gray bar for reference. (B) Sequence alignment of obtained mutants compared to wild-type ACP1 and ACP2. Randomized positions are indicated with a yellow asterisk. (C) Turnover rates of wild-type and chimeric bimodular PKSs harboring the mutations enriched in the ACP1-Lib1 biopanning experiments. All bimodular PKS consisted of LM, M1, and Module“X”-TE. Either wild-type DEBS M1 was used as the first module (M1) or one of the six mutants obtained through the first directed evolution experiments (M1-Lib1-Mut“X”). Initial rate data were obtained at individual PKS protein concentrations of 4 μM and nonlimiting concentrations of propionyl-CoA, methylmalonyl-CoA, and NADPH. Measurements were performed in triplicate from one batch per construct M1-Lib1-Mut“X”, and mean and standard deviation are given.