Figure 2.
Barcode Cycle Sequencing (BCS) components: a strategy for converting amino acids into DNA barcodes directly on a next-generation sequencing chip
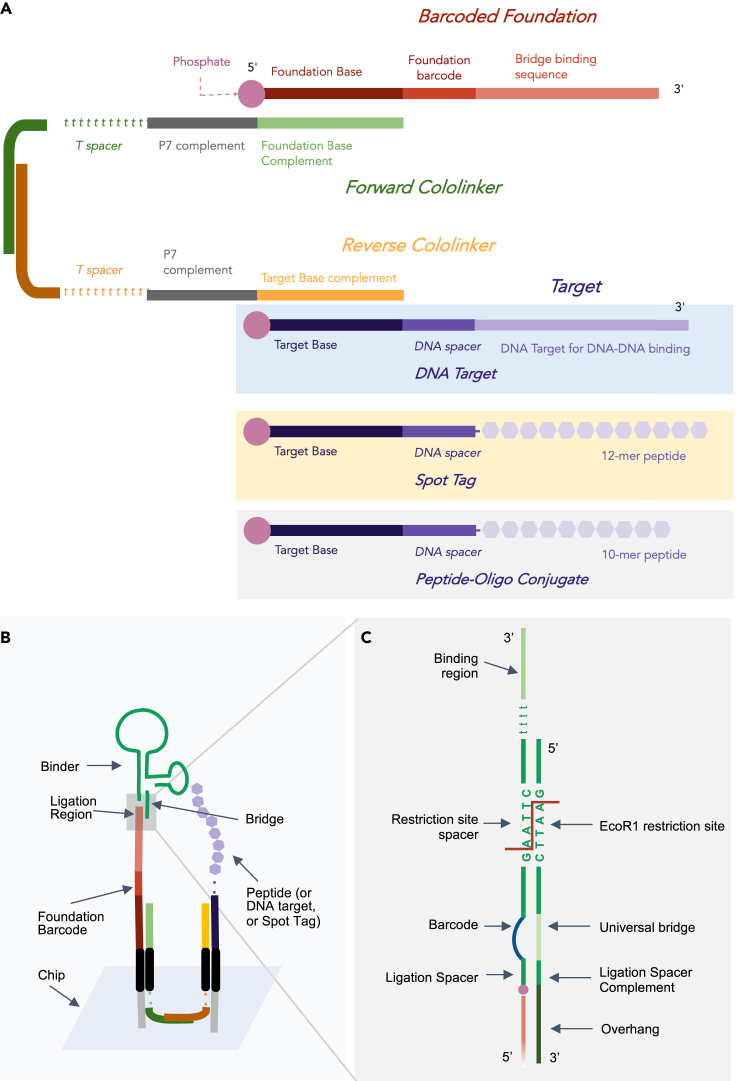
(A) Foundations are assembled off-chip. Two cololinkers that are partially complementary to each other and complementary to P7 adapters are used to link a barcoded foundation with the oligo-tethered target to be sequenced. Experimentally, this target may consist of DNA, a Spot-Tag with residues, or a 10-mer peptide.
(B) Targets and foundation barcodes are deposited in close proximity on the sequencing chip by ligating the target and foundation barcode to proximal P7 adapters on the sequencing chip. The cololinkers allow the foundation barcode and target to localize to adapters in close proximity. Cololinkers are washed away prior to binding events and no longer present.
(C) Depicts the gray region of B in detail. The 5′ end of the oligo portion of each binder contains a restriction site spacer, which is hybridized to a complementary universal bridge. The bridge provides a double-stranded substrate that the restriction enzyme can act upon. Full sequences can be found in Table S2 and molecular details in Figure S2.