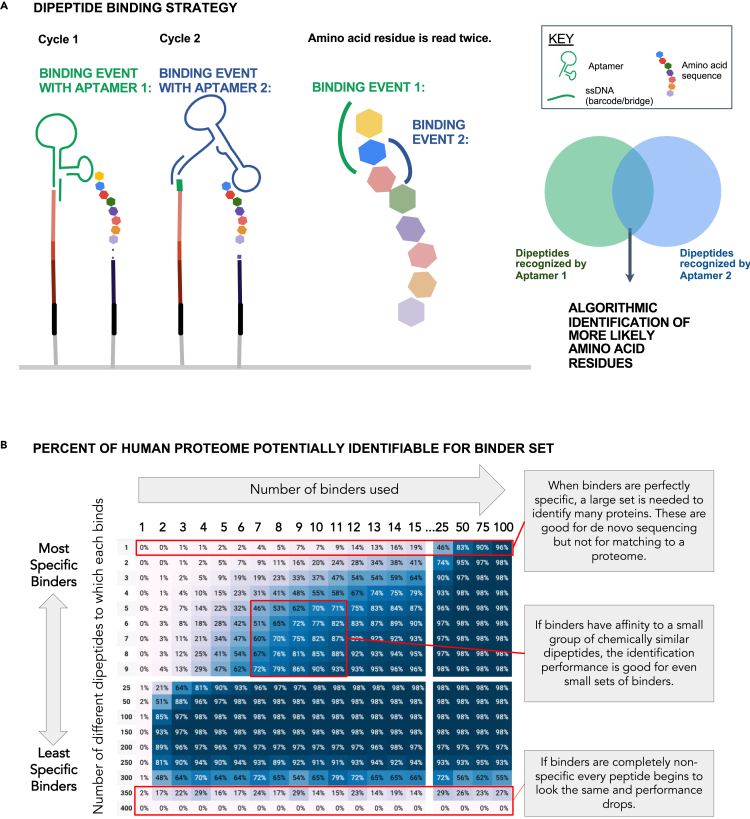
Figure 7.
A binder set for the algorithmic identification and coverage of the human proteome
(A) A dipeptide aptamer binder provides putative identities for the two N-terminal amino acids. As each round of Edman degradation removes only one amino acid, each amino acid (except the original N-terminal amino acid) is exposed to two rounds of aptamer binding, enabling algorithmic identification of individual residues based on overlap between likely candidates identified across two rounds.
(B) A simulation predicts that a small set of semi-selective binders offers significant coverage of the human proteome. Binder sets of various sizes and selectivity were evaluated to see what percent of the proteome could be identified. In the simulation, each binder in a set binds to a sample of the 400 possible dipeptides (20 possibilities for two N-terminal amino acids). A protein is identified if the barcode series for a sequenced fragment is unique. See the text for details of the simulation. For each actual binding set, the real-world performance would be contingent on the set-specific binding characteristics (or parameters).