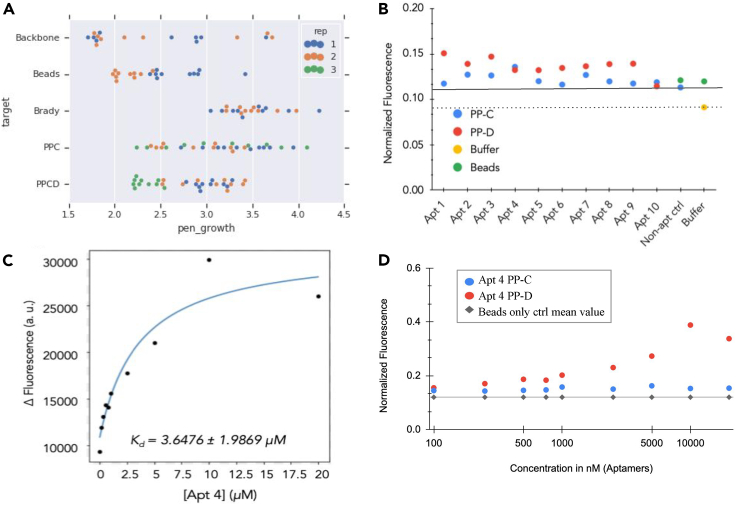
Figure 9.
Aptamer 4 Kd and backbone dependence
(A) Top ten sequences for each selection for each target. Two selections each were performed for Backbone, Beads, and Bradykinin. Three selections were performed for PPC and PPCD. High enrichment (>3, equivalent to 1,000-fold) was seen for 4 sequences for Backbone, 1 sequence for beads, all of the top 10 sequences (total 20) for bradykinin, 18/30 sequences for PPC, and 11/30 sequences for PP-CD.
(B) Results of a single point binding assay for 10 potential aptamer candidates. Binding, indicated by fluorescent signal (y axis), was measured for 10 aptamers at 100 nM. Apt 4 shows higher binding than the controls (non-aptamer [straight line] and buffer [dotted line]) for target PP-C. Apt 1,2,3,4,7,8,9 show higher binding than controls for PP-D. Data were normalized to the positive control (FAM conjugated directly to beads).
(C) Binding curve for Apt 4 binding to 100 nM PP-D plotted here. Data were fitted via the “fit_hyperbola” function in the biofits library (https://github.com/jimrybarski/biofits). Apt 4's Kd was found to be 3.65 μM (±1.99).
(D) Binding curve for Apt 4 for targets PP-C and PP-D for 100 nM to 2.5 mM concentrations. Apt 4 shows saturation binding against PP-D and no binding against PP-C. Data were normalized to the positive control (FAM conjugated directly to beads).