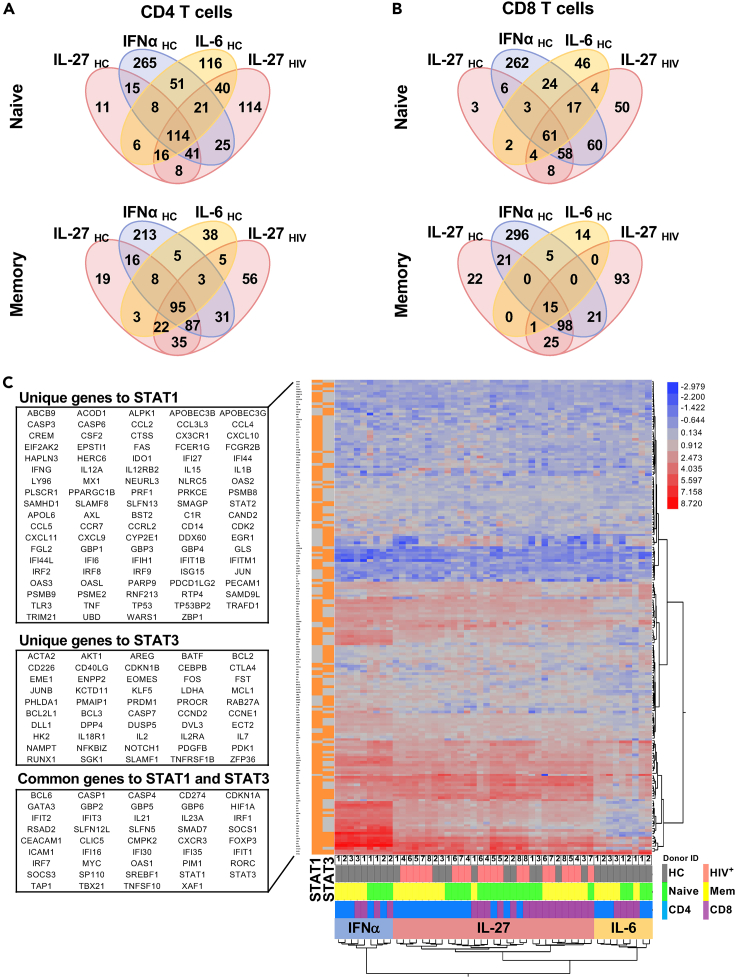
Figure 2.
IL-27 signaling induced a cluster of STAT1-dependent genes
RNAseq analysis of sorted naive (CD45RA+CD27+) and memory (CD45RA−CD27+) T cells from PLWH (HIV+, n = 5) and healthy controls (HC, n = 3) stimulated in vitro with 100 ng/mL of rhIL-27, and cells from healthy controls stimulated with 100 U/mL of rhIFN⍺ and 100 ng/mL of rhIL-6 for 90 min as controls.
(A) Venn diagrams represent overlapped differentially expressed gene transcripts (DEGs) regulated by IL-27 in CD4 T cell subsets from PLWH and HC compared to the DEGs induced by IFN⍺ and IL-6 stimulation in T cells from healthy controls.
(B) Venn diagrams represent overlapped DEGs regulated by IL-27 in CD8 T cell subsets from PLWH and HC compared to the DEGs induced by IFN⍺ and IL-6 stimulation in T cell from healthy controls.
(C) Ingenuity Pathway Analysis (IPA) was used to subset the RNA-seq data to include only target genes predicted by IPA to be regulated via STAT1 and/or STAT3 activity, as indicated in the left column, orange for genes downstream, gray for genes not downstream of the corresponding upstream regulator. The 2-way clustered heatmap of gene expression (log2 of stimulated/unstimulated ratio) for IL-27, IFNα and IL-6 stimulated T cell subsets. The color bar indicates: HC group (gray), HIV group (pink), CD4 T cells (blue), CD8 T cells (purple), naive subset (green), Memory (Mem, yellow). The lists (DEGs) were selected based on >2-fold change (| Log2 FC | > 1) and FDR <0.05.