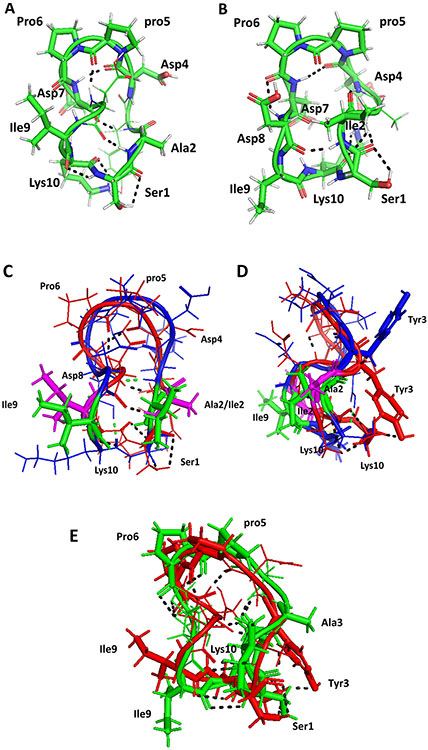
FIGURE 3.
Proposed model for the structures of A) AS2 and B) AS3. Structures were generated using NMR-restrained molecular dynamics simulations followed by energy minimization. Amino acid residues that are in β-turns in the structure are shown by sticks and are labeled with three-letter codes for amino acids. C) Overlay of structures of peptide 6 (backbone blue) and AS2 (backbone red). Ile2 and Ile9 from peptide 6 are shown as sticks in green. Ala2 and Ile9 from AS2 are shown in magenta. There is a slight change in the backbone structure of AS2 compared to peptide 6. D) Side view of overlay of peptide 6 and AS2. E) Overlay of structures of peptides AS2 (red) and AS3 (green). Replacement of Tyr 3 by Ala changes the overall backbone structure slightly, but the bulky side chain of Tyr is important in interaction with CD58 protein. Hydrogen bonds are shown by dashed lines. pro5 represents the D amino acid proline.