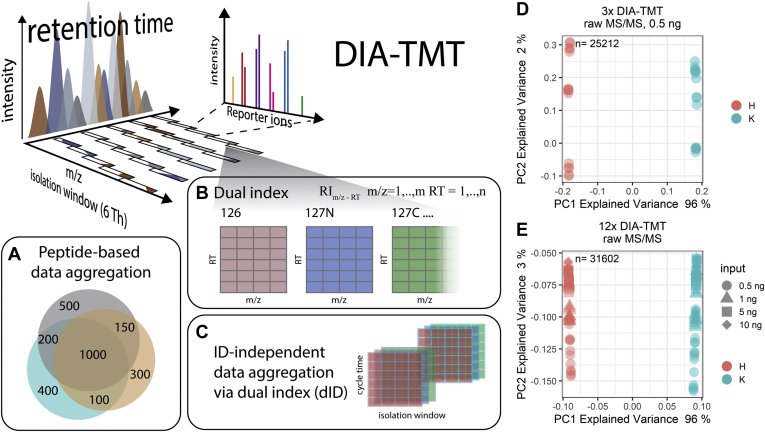
Fig. 1.
ID-independent DIA-TMT analysis creates cell type specific clusters.A, peptide-based data aggregation of DIA-TMT results in a decrease overlap between replicates without computational generation of quantitative data. B, all MS/MS scans from the DIA-TMT files are indexed using the dual indexing (dID) = [RT1,2,…,m × m/z1,2,…,n] according to the RT (RT 1, 2, …, m) and central mass (m/z 1, 2, …, n). In conjunction with the quantitative RI values, a grid-like 3D map or proteome signature is created, which is used for (C) ID-independent data aggregation theoretically resulting in a complete overlap of all MS/MS scans. PCA of (D) three DIA-TMT runs (30 multiplexed samples) at 0.5 ng total peptide input or (E) 12 DIA-TMT runs (120 multiplexed samples) at four peptide inputs (0.5, 1, 5, 10 ng) ID-independently aggregated. Samples are colored according to channel loadings and the respective peptide inputs are indicated with different symbols. H = HeLa cells, K = K562 cells. n, number of MSMS scans included in PCA.