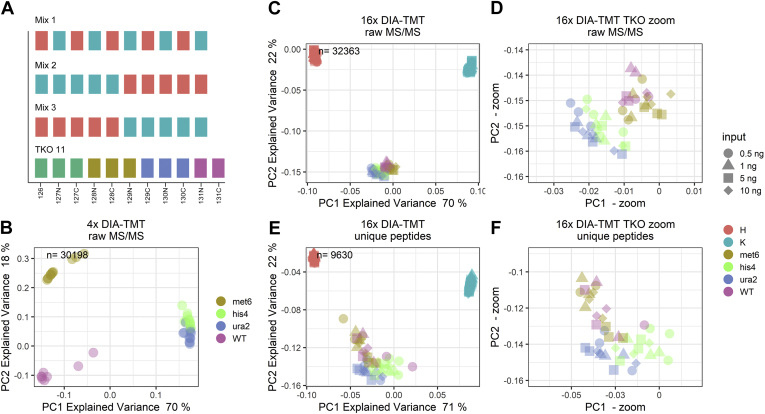
Fig. 2.
Proteome signatures are input, batch, and species-independent across large sample cohorts.A, overview of isobaric labeled mixes. PCA of (B) four and (C) 16 DIA-TMT runs ID-independently aggregated at four peptide inputs (0.5, 1, 5, 10 ng). D, zoom into TKO11 cluster displayed in panel C. E, PCA of 16 DIA-TMT runs at four peptide inputs (0.5, 1, 5, 10 ng) with standard peptide-based aggregation and (F) zoom into TKO11 cluster displayed in panel E. Samples are colored according to channel loadings and the respective peptide input is indicated with different symbols. H = HeLa cells, K = K562 cells and the respective TKO11 strains (i.e., WT, knockouts: met6, his4, ura2). n, number of unique peptides included in PCA.