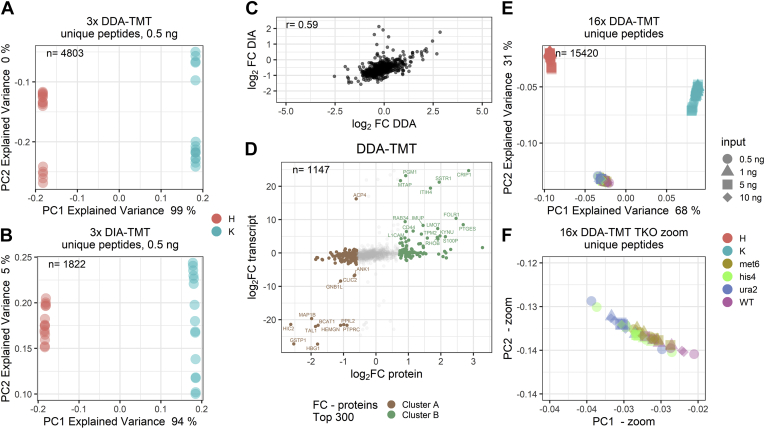
Fig. 4.
Identification-independent data aggregation allows for the analysis of closely related cell types. PCA of three analytical runs with standard peptide-based data aggregation at 0.5 ng total peptide input and missing value imputation of (A) DDA or (B) DIA acquisition schemes. n = number of unique peptides included in PCA. C, correlation of the average log2 fold change of proteins identified in DDA and DIA experiments displayed in Figures 3A and 4C. r = Pearson correlation estimate. D, intersection of transcriptome (FPKM) with DDA proteome data with top 300 proteins colored according to cluster contributions and top 60 proteins are labeled. n = protein identifications across all analytical runs after missing value imputation. PCA of 16 DDA runs based on peptide-based data aggregation with missing value imputation runs at four peptide inputs (0.5, 1, 5, 10 ng). F, zoom into TKO11 cluster displayed in panel E. Samples are colored according to channel loadings and the respective peptide input is indicated with different symbols. H = HeLa cells, K = K562 cells and the respective TKO11 strains (i.e., WT, knockouts: met6, his4, ura2). n, number of unique peptides included in PCA.