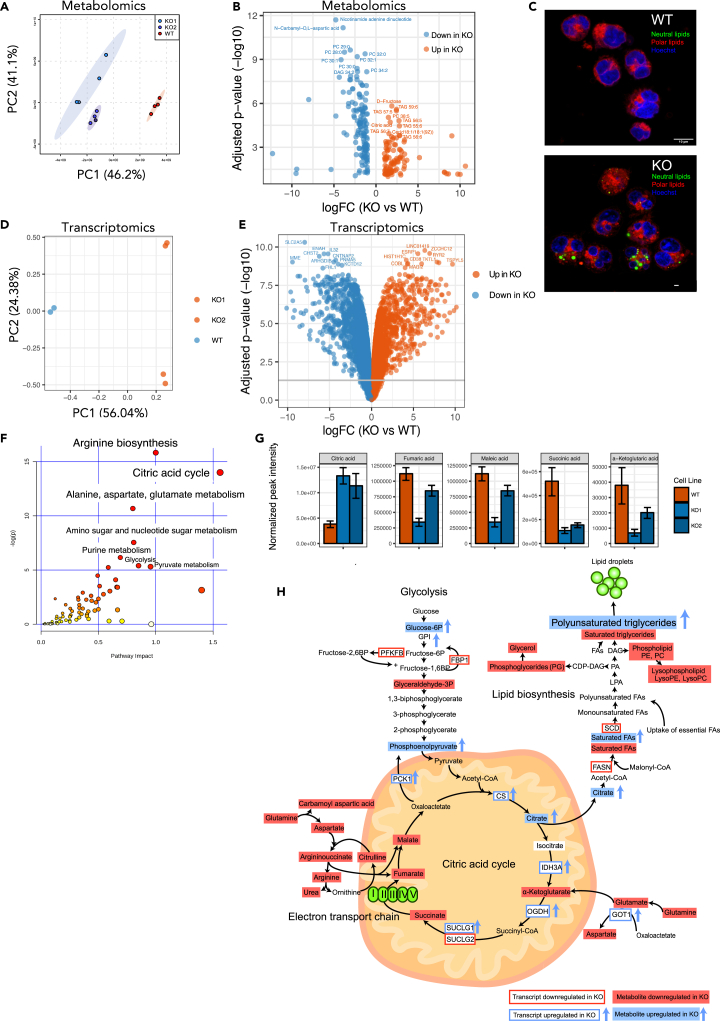
Figure 4.
Loss of IL-32 leads to perturbations in metabolic pathways
(A) PCA plot of metabolomes from two clones of INA-6 KO cells and WT mock cells.
(B) Volcano plot showing significant different metabolites (p <0.05) between KO cells and WT mock cells (metabolite expression from replicates from two KO clones were merged) See also Table S1. Significance was determined by two-sided Student's t test using MetaboAnalyst 4.0 software.
(C) Representative image of lipid droplets in INA-6 IL-32 KO and WT mock cells, stained with Nile Red and Hoechst. Polar lipids (red) were excited at 590 nm (600–700 nm) and neutral lipids (green) at 488 nm (500–580 nm). Confocal imaging was performed with a Leica TCS SP8 STED 3X, using a 63 × 1.4 (oil) objective and LAS X software. Scale bar: 10 μM. See Figure S4 for overview images.
(D) Two INA-6 KO cell lines and WT mock cells were subjected to RNA sequencing, and the PCA plot shows the overall differences in gene expression between KO cells and WT mock cells.
(E) Volcano plot showing the most significantly upregulated and downregulated genes in INA-6 KO cells (2 clones) versus WT mock cells. Statistical significance analyzed by Linear Models for Microarray Analysis (limma) in R with Benjamini-Hochberg-adjusted p values. See also Table S2 for complete gene list.
(F) Joint pathway analysis (SMPDB pathways, MetaboAnalyst 4.0) of transcriptomic and metabolomic data from 2 INA-6 IL-32 KO clones and WT mock cells. The inverse logged p-value of the different pathways is shown on the y-axis, and the size and color on the dots (increased size and increasingly red) correspond to the increased inverse log p-value. Significance was determined by two-sided Student's t test using MetaboAnalyst 4.0 software. The joint pathway analysis is based on metabolites in Table S1 and genes (fold change >0.5 or < −0.5 and adjusted p value <0.05) in Table S2.
(G) Significantly (p <0.05) altered citric acid cycle intermediates in two KO clones (KO1, KO2) versus WT mock cells (See also Table S1). Data are presented as mean peak intensity ± SD of 4 replicates.
(H) Illustration of significantly differentially expressed genes and metabolites from the most enriched pathways in the joint pathway enrichment analysis in (F) Significance was determined by two-sided Student's t test using MetaboAnalyst 4.0 software.