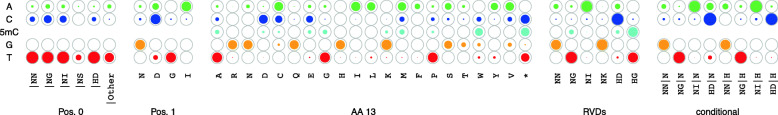
Fig. 1.
Parameters of the EpiTALE model. Parameters are represented by circles that are arranged in rows according to the bound nucleotide including 5mC. Column labels indicate the RVD, 13th AA, or combination of RVD and previous 12th AA that a parameter depends on. The outer circles are filled by a coloured circle proportional to the corresponding specificity parameters, where different colours are used for the four nucleotides and 5mC, respectively. For instance, the RVD ’HD’ has a strong preference for C, while A, 5mC and T are bound in decreasing preference, and G is a clear mismatch for ’HD’. There are no separate parameters for methylated cytosines for the sub-model at position 0, position 1 and the “conditional” sub-model, as there are no sufficient data available yet