Extended Data Fig. 4|. The structure of inactive S. pombe Arp2/3 complex is nearly identical to other inactive Arp2/3 complex structures.
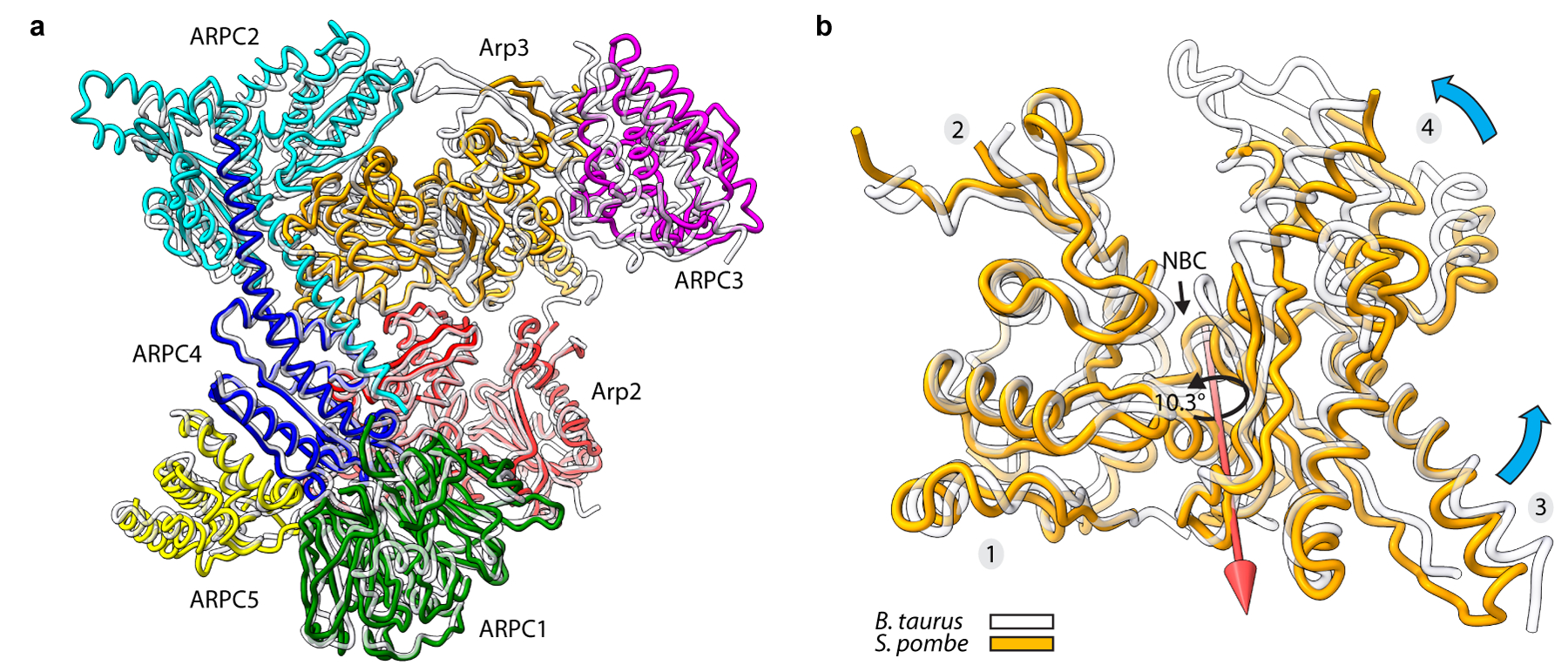
a, Structural superposition of inactive S. pombe Arp2/3 complex (colored subunits) from the cryo-EM reconstruction presented here with inactive Bos taurus Arp2/3 complex (semi-transparent gray subunits, PDB:4jd218). Structures were superposed using Arp3’s subdomains 1 and 2, Arp2, ARPC1, ARPC2 and ARPC4. b, Structural superposition showing that Arp3 from the S. pombe inactive Arp2/3 complex structure shows a more open nucleotide binding cleft than other ATP-bound structures. Subdomains 1 and 2 of inactive S. pombe Arp3 (orange) were superposed on inactive BtArp3 (transparent gray, 4jd2). Cyan arrow shows the direction of rotation of subdomains 3 and 4 toward subdomains 1 and 2 during cleft closure. Red arrow shows the axis of rotation of cleft closure. A rotation of 10.3º about this axis is required to close the S. pombe nucleotide cleft to the same extent as the BtArp2/3 structure (PDB 4jd2). The open cleft in the inactive S. pombe Arp2/3 complex is the major structural difference between the two structures.
