Extended Data Fig. 2|. Single-particle cryo-EM data processing workflow for 3D reconstructions of activated and inactive Arp2/3 complex.
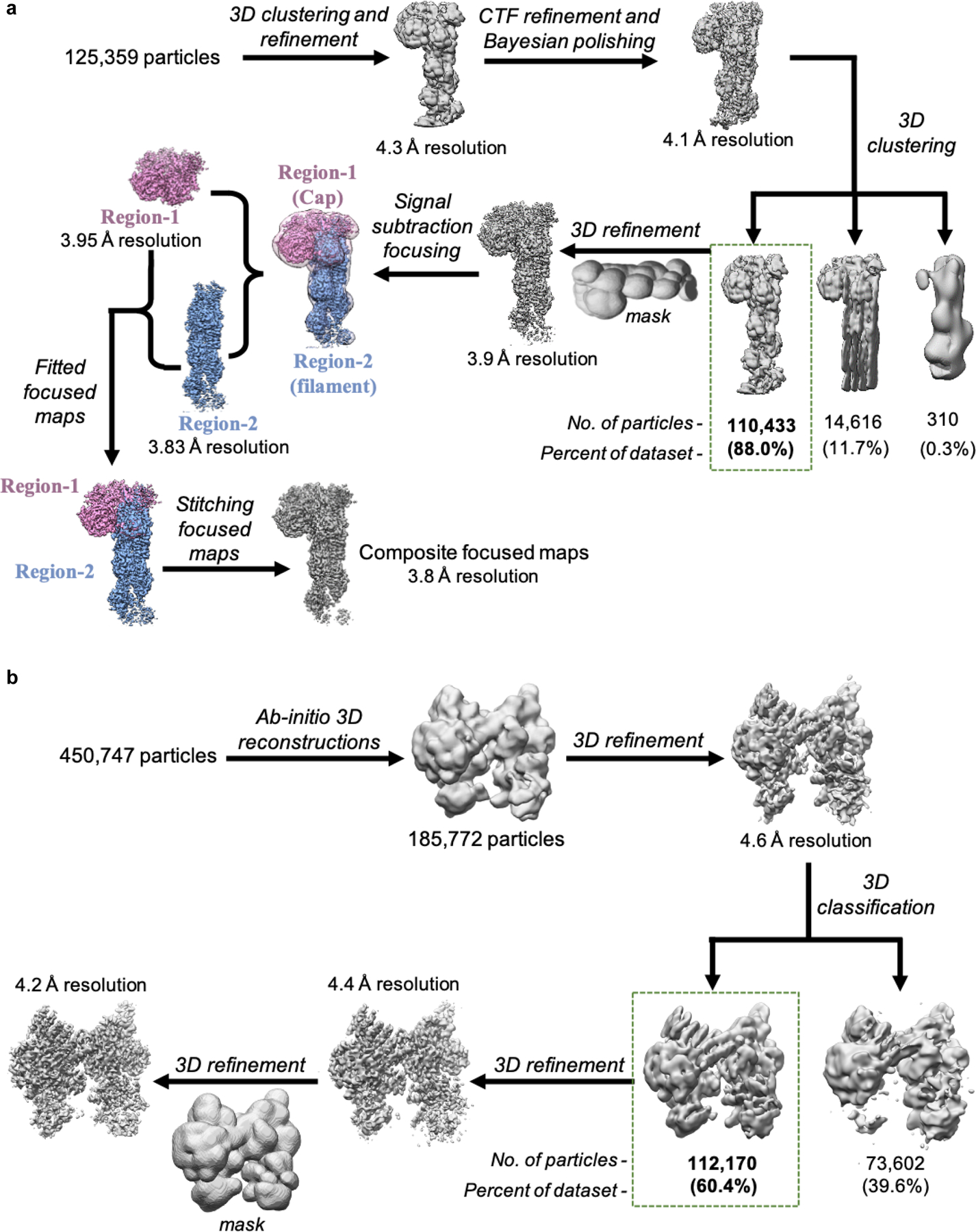
a, Data processing schematic for Dip1-Arp2/3-actin-filament complex. Particle stack was initially binned by factor of four and subjected to multiple rounds of 2D and 3D classification to discard particles that did not correspond to the complex. The cleaned particle stack was re-extracted without binning and further subjected to downstream 3D processing. All 3D classifications were performed without image alignment (referred to as clustering). Final composite map was generated by combining focused maps corresponding to the Dip1-Arp2/3 cap (Region-1, colored purple) and the Arp-actin filament (Region-2, colored blue). b, Schematic for processing of Arp2/3 complex (inactive state) cryo-EM data. Particles extracted from all micrographs (tilted and non-tilted) were cleaned by multiple rounds of 2D classification and then subjected to multiple ab initio 3D reconstructions. Particles corresponding to intact Arp2/3 complex were further subjected to downstream data processing that lead to a 4.2 Å 3D reconstruction of Arp2/3 complex in inactive state.
