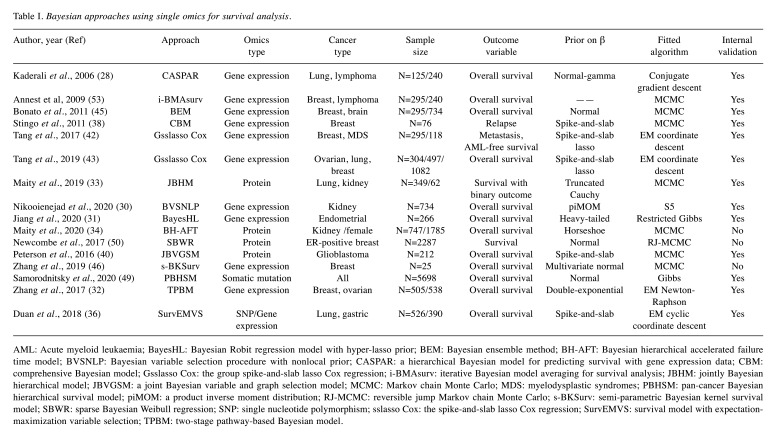
Table I. Bayesian approaches using single omics for survival analysis.
AML: Acute myeloid leukaemia; BayesHL: Bayesian Robit regression model with hyper-lasso prior; BEM: Bayesian ensemble method; BH-AFT: Bayesian hierarchical accelerated failure time model; BVSNLP: Bayesian variable selection procedure with nonlocal prior; CASPAR: a hierarchical Bayesian model for predicting survival with gene expression data; CBM: comprehensive Bayesian model; Gsslasso Cox: the group spike-and-slab lasso Cox regression; i-BMAsurv: iterative Bayesian model averaging for survival analysis; JBHM: jointly Bayesian hierarchical model; JBVGSM: a joint Bayesian variable and graph selection model; MCMC: Markov chain Monte Carlo; MDS: myelodysplastic syndromes; PBHSM: pan-cancer Bayesian hierarchical survival model; piMOM: a product inverse moment distribution; RJ-MCMC: reversible jump Markov chain Monte Carlo; s-BKSurv: semi-parametric Bayesian kernel survival model; SBWR: sparse Bayesian Weibull regression; SNP: single nucleotide polymorphism; sslasso Cox: the spike-and-slab lasso Cox regression; SurvEMVS: survival model with expectation-maximization variable selection; TPBM: two-stage pathway-based Bayesian model.