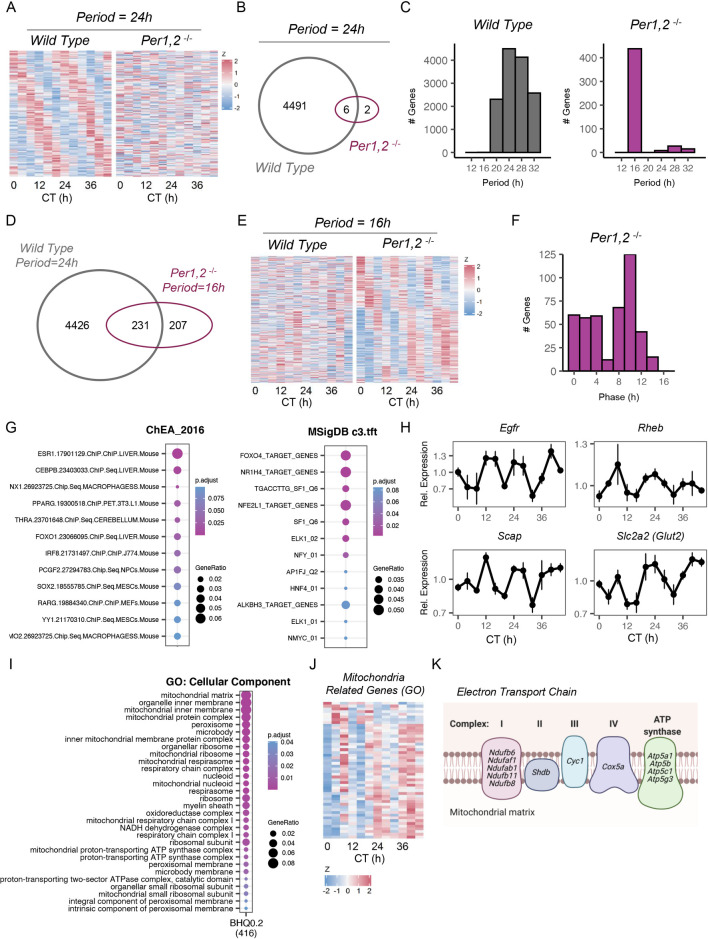
Fig 3. Liver gene expression cycles with ultradian periodicity in Per1,2−/− mice under free-running conditions.
(A) Heatmap of expression profiles of genes that were rhythmic (q < 0.2, JTK_CYCLE analysis) in WT with an about 24-hour period and their corresponding profiles in Per1,2−/− mice. Data are presented as z-scores of the average expression in each CT. (B) Venn diagrams representing the overlap between 24-hour rhythmic genes in WT or Per1,2−/− mice. (C) Periodograms of the transcriptome in WT or Per1,2−/− mice. (D) Venn diagrams representing the overlap between 24-hour rhythmic genes in WT or 16-hour rhythmic genes in Per1,2−/− mice. (E) Heatmap of expression profiles of the rhythmic genes in Per1,2−/− mice with an about 16-hour period and their corresponding profiles in WT. Data are presented as z-scores of the average expression in each CT. (F) Histogram representing the distribution of phases of 16-hour rhythmic genes in Per1,2−/− mice. (G) Enrichment analysis of rhythmic genes of 16-hour rhythmic genes in Per1,2−/− mice based on ChEA dataset (left) and MSigDB C3:TFT collection (right) (p < 0.1, overrepresentation test). (H) Daily profiles of selected 16-hour rhythmic genes in Per1,2−/− mice. (I) GO cellular compartment enrichment analysis of 16-hour rhythmic genes in Per1,2−/− mice (p < 0.05, overrepresentation test). (J) Heatmap of expression profiles of mitochondria related genes that were rhythmic with about 16-hour period in Per1,2−/− mice. Data are presented as z-scores of the average expression in each CT. (K) Graphic representation of mitochondrial electron transport chain complexes, highlighting related genes that were rhythmic with about 16-hour period in Per1,2−/− mice. See also S1 and S2 Tables. CT, circadian time; GO, Gene Ontology; WT, wild-type.