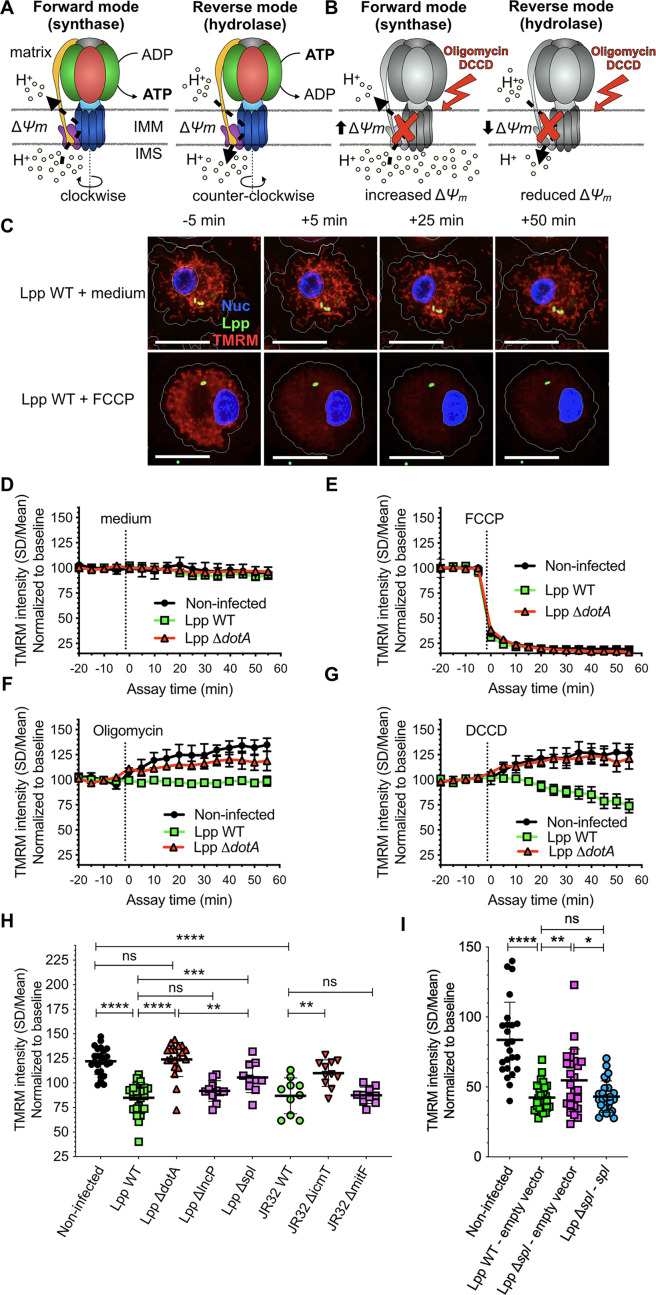
Figure 2. The mitochondrial FOF1-ATPase works in the ‘reverse mode’ during infection of human monocyte-derived macrophages (hMDMs) by L. pneumophila.
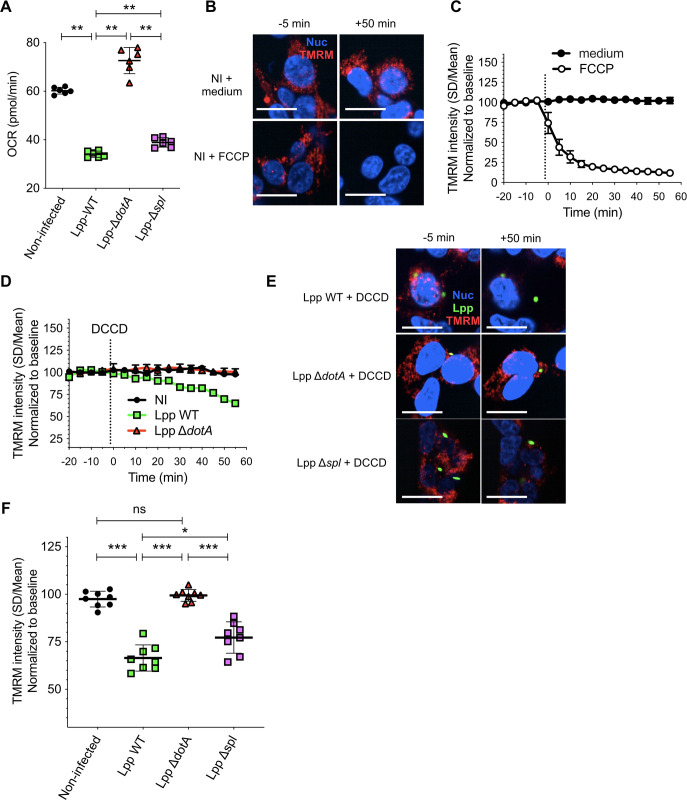
(A) In the ‘forward mode’ of the mitochondrial ATPase, the Δψm generated by the electron transport chain is used by the FOF1-ATPase to synthesize ATP. The ‘reverse mode’ of the FOF1-ATPase leads to ATP hydrolysis to pump H+ to the intermembrane space (IMS). IMM, inner mitochondrial membrane. (B) When the FOF1-ATPase is inhibited by oligomycin or dicyclohexylcarbodiimide (DCCD), an increase in Δψm indicates that the ATPase was working in the ‘forward mode’ (H+ accumulate in the IMS), while a decrease in Δψm indicates functioning in the ‘reverse mode’ (H+ cannot be translocated to the IMS by the FOF1-ATPase to sustain the Δψm). (C) hMDMs were infected with GFP-expressing bacteria (green) or left uninfected (noninfected). At 5.5 hr post-infection (hpi), cells were labeled with Hoechst to identify the cell nucleus (Nuc, blue) and tetramethylrhodamine methyl ester (TMRM) (red) to quantify Δψm. At 6 hpi, addition of medium (no changes) or FCCP (complete depolarization) was used as controls. Representative confocal images of Lpp-WT-infected hMDMs (6 hpi) at 5 min before the addition of medium (top) or FCCP (bottom), and at 5, 25, and 50 min after addition of medium or FCCP. Bar: 20 μm. (D) Quantification of (C) before (baseline) and after the addition of medium. Each dot represents mean ± SD of three independent experiments with a total of eight replicates. (E) Same as (D) but FCCP was added. (F) Same as (D) but oligomycin was added. (G) Same as (D) but DCCD was added. (H) Same as (C) but infection was performed with Lpp-WT, Lpp-ΔdotA, Lpp-ΔlncP, Lpp-Δspl, L. pneumophila strain Philadelphia JR32 (JR32)-WT, JR32-ΔicmT or JR32-ΔmitF. TMRM values (SD/mean) at 50 min after DCCD addition are shown. Data from a minimum of three experiments per strain with 10 or more replicates per strain. (I) hMDMs were infected with GFP-expressing bacteria or left uninfected (noninfected). At 5.5 hpi, cells were labeled with Hoechst to identify the cell nucleus and TMRM to quantify Δψm. At 6 hpi, addition of DCCD revealed whether the FOF1-ATPase works in the ‘reverse mode.’ Infection was performed with Lpp-WT expressing empty pBCKS vector (empty vector), Lpp-Δspl-expressing empty vector and Lpp-Δspl-expressing pBCKS-spl vector, which express LpSpl (complemented strain, Lpp-Δspl:: spl). Data from three donors are shown. Each dot represents a replicate. *p<0.1; **p<0.01; ***p<0.001; ****p<0.00001; ns, nonsignificant (Mann–Whitney U test).