Abstract
PURPOSE
To examine the association of gain-of-function (GOF) and non–gain-of-function (non-GOF) TP53 mutations with prognosis of metastatic right-sided (RCC) versus left-sided colorectal cancer (LCC).
METHODS
This cohort study included patients with metastatic colorectal cancer (CRC) who had next-generation sequencing performed from November 2017 to January 2021. We defined R175H, R248W, R248Q, R249S, R273H, R273L, and R282W as GOF and all other mutp53 as non-GOF. We used Cox regression modeling to examine the association between GOF and non-GOF mutp53 and overall survival (OS), adjusting for age, sex, ethnicity, performance status, Charlson comorbidity index and receipt of chemotherapy.
RESULTS
Of total 1,043 patients, 735 had tumors with mutp53 and 308 had wild-type p53 (wtp53). GOF was associated with worse OS than non-GOF mutp53 only in LCC (hazard ratio [HR] = 1.66 [95% CI, 1.20 to 2.29]), but not in RCC (HR = 0.79 [95% CI, 0.49 to 1.26]). Importantly, RCC was associated with worse OS than LCC only in the subset of patients whose CRC carried non-GOF (HR = 1.76 [95% CI, 1.30 to 2.39]), but not GOF mutp53 (HR = 0.92 [95% CI, 0.55 to 1.53]) or wtp53 (HR = 0.88 [95% CI, 0.60 to 1.28]). These associations were largely unchanged after also adjusting for RAS, BRAF, and PIK3CA mutations, and microsatellite instability-high.
CONCLUSION
Poorer survival of patients with metastatic RCC versus LCC appeared to be restricted to the subset with non-GOF mutp53, whereas GOF versus non-GOF mutp53 was associated with poorer survival only among patients with LCC. This approach of collectively classifying mutp53 into GOF and non-GOF provides new insight for prognostic stratification and for understanding the mechanism of sidedness-dependent prognosis. If confirmed, future CRC clinical trials may benefit from incorporating this approach.
INTRODUCTION
TP53 mutation is present in more than 50% of colorectal cancer (CRC),1-5 and is implicated in the majority of Li-Fraumeni syndrome cases, a hereditary cancer syndrome associated with early onset of leukemia and solid tumors.6,7 p53 regulates numerous biologic processes including DNA repair, cell cycle arrest, apoptosis, cell senescence, metabolic pathways, and others primarily by modulating the expression of its target genes.4,8-11
CONTEXT
Key Objective
The prognostic significance of p53 mutations in colorectal cancer (CRC) has not been established. This study investigates whether p53 mutations, when classified into gain-of-function (GOF) and non–gain-of-function (non-GOF) on the basis of review of current literature, could stratify the prognosis of right-sided versus left-sided metastatic CRC.
Knowledge Generated
Our study reveals that non-GOF p53 mutations were associated with worse prognosis of metastatic right-sided versus left-sided colorectal cancer, while GOF versus non-GOF p53 mutations were associated with worse prognosis in the left-sided but not right-sided colorectal cancer.
Relevance
Our findings suggest that different p53 mutations may differentially affect survival of patients with metastatic CRC on the basis of anatomic location and shed new light on the potential oncogenic mechanisms of p53 mutations in CRC. GOF versus non-GOF p53 mutations may collectively serve as useful biomarkers in guiding clinical practice with prognostic stratification and in the design of future CRC clinical trials.
The spectrum of p53 mutation (mutp53) is broad and varies in various malignancies, including missense mutation, nonsense mutation, deletion, frameshift, insertion, etc. Approximately 80% of these mutations occur in the DNA binding domain (DBD), most of which are missense mutations.3,10,12,13 Approximately 30% of the mutations cluster within six mutation hotspots in the DBD: R175, G245, R248, R249, R273, and R282.3,12,14 Several of these mutp53 have been shown to exhibit gain-of-function (GOF) properties, possessing novel functions that are not part of the wild-type p53 protein (wtp53).3,12,15,16 The concept of GOF was first demonstrated by Dittmer et al who ectopically expressed mutp53 R175H and R273H in p53-null cells and showed enhanced tumorigenic potential in nude mice, higher efficiency in forming colonies in soft agar, and increased expression of drug-resistant genes.3,12,17 This was consistent with the observation of patients with Li-Fraumeni syndrome whose onset of malignancies was significantly earlier when a missense mutation such as R175H was responsible compared with TP53 deletion that causes the loss of function.7,18 This observation could be replicated in knock-in mice carrying R175H, R273H, or R248Q that developed epithelial cancer with more rapid progression and shorter survival compared with the knock-out mice (with deletion of both TP53 alleles) that primarily develop lymphoma.19-21 Numerous experimental studies have found that GOF mutants increase cell invasion and proliferation, chemoresistance, colony formation, genomic instability, angiogenesis, etc.3,12,15 GOF mutants can bind to novel proteins, form new interactions, enhance the signaling of receptor tyrosine kinases, and drive the expression of NF-kB, etc.3,12,15,22-26
In addition to the established molecular features (RAS, BRAF, and PIK3CA mutations, and microsatellite instability-high [MSI-high]) that are closely related to the survival of metastatic CRC, anatomic location (sidedness) of the primary tumor has emerged in recent years as an important prognostic factor.27,28 Many studies have shown that metastatic right-sided CRC (proximal, RCC) is associated with worse survival than metastatic left-sided CRC (distal, LCC), including findings from several randomized clinical trials and a large meta-analysis.29-32 The poorer survival of patients with RCC appears to be independent of the molecular features; however, its underlying mechanism remains unclear.27,28
Despite many lines of experimental evidence for p53 GOF in cell lines and animals, the data in human malignancies are limited. Our goal was to investigate the p53 GOF concept in human malignancy using the next-generation sequencing (NGS) data within Kaiser Permanente Northern California (KPNC) and to explore the relations between the sidedness of metastatic CRC, p53 mutation, and overall survival (OS).
METHODS
Study Population
Our data set includes KPNC patients with stage IV CRC with StrataNGS (Ann Arbor, MI) performed from November 2017 to January 2021. Patient data on demographics, Charlson comorbidity index (CCI), performance status (PS), and lines of chemotherapy received were retrieved from the electronic medical record (Epic) and cancer registry database. CCI was based on the 12 month-period before the date of diagnosis of metastatic CRC. This study was approved by the KPNC institutional review board with waiver of consent.
StrataNGS
StrataNGS of advanced malignancies began in November 2017 in KPNC and initially included approximately 90 commonly mutated genes including TP53, KRAS, NRAS, BRAF, PIK3CA, and MSI, expanded to include other genes in August 2019, and is currently a 429-gene, pan-solid tumor, NGS assay for formalin-fixed, paraffin-embedded tumor tissue, performed on coisolated DNA and RNA.33
Definition of GOF and Non-GOF p53 Mutation
There are no common established criteria for all the GOF p53 mutations in the literature. We defined the GOF and non-GOF in our data set on the basis of our literature review. There were 299 different individual p53 mutations identified in our data set. We focused on the six mutation hotspots in the DBD to examine the impact of mutp53 GOF: R175H, G245S, R248Q and R248W, R249S, R273H and R273L, and R282W.3,12 Although a few other mutp53 have been shown to possess some GOF properties, the strength of evidence does not appear strong.3,12 We further excluded G245S because the evidence was weak for it to be considered a GOF. For example, in a mouse knock-in experiment, G245S knock-in mice exhibited later onset of tumors and longer survival compared with the R248Q knock-in, similar to the mice that carried TP53 deletion.20 Also, patients with Li-Fraumeni syndrome carrying a germline G245S mutation had later onset of tumors compared with R175, R248, and R282 mutations.19 Multiple GOF properties have been demonstrated with mutp53 R175H, R248Q, R248W, R249S, R273H, R273L, and R282W, including increased invasion, cell proliferation, chemoresistance, and tumor growth in xenograft.3,12,14,15 We identified a total of 204 patients with GOF in our cohort, including 83 patients with an R175H, 39 with an R248Q, 21 with an R248W, 4 with an R249S, 35 with an R273H, one with R273L, and 21 with an R282W. We classified all other mutp53 as non-GOF.
Definition of RCC and LCC
RCC was defined as colon cancer of ascending and transverse colon, and LCC defined as CRC from splenic flexure to rectum.
Statistical Analysis
OS was measured from the time of diagnosis of stage IV CRC to the time of death or end of follow-up (April 16, 2021, last OS data pull), whichever came first. Patients who were still alive at the end of follow-up were censored. Pearson's chi-squared test was used to determine statistical difference on demographics and distribution of p53, RAS, BRAF, and PI3K mutations, and MSI-high. We used the one-way analysis of variance test to assess differences in continuous variables. We used Kaplan-Meier plot to perform unadjusted OS analysis and estimate median OS. The number of patients at risk under the OS curves reflects delayed entry into the cohort at the time of receipt of NGS results (ie, left truncation, with study entry ranging from 0 to 9 years after diagnosis, median 4.0 months).34 The prespecified alpha error was set at .05 in two-tailed test. To adjust for multiple testing, we controlled the false discovery rate at the level α = .05 and determined statistical significance by using the Benjamini-Hochberg procedure.35 Cox proportional hazards regression models were used to estimate adjusted hazard ratios (HRs) and 95% CIs for the association between p53 mutations and OS, adjusted for covariates. Time since diagnosis of stage IV CRC was the time scale used in the regression models, allowing for left truncation. In our primary analysis, covariates included in our main regression models were age (continuous), sex (male and female), ethnicity (non-Hispanic White, Black, Asian, Hispanic, and other or unknown), PS (0-1, 2-3), CCI (continuous), and chemotherapy received (yes or no). In our secondary models, we also included the molecular features (RAS [yes or no], BRAF [yes or no], and PIK3CA [yes or no] mutations, and MSI-high [yes or no]). PS was the only variable with missing or unknown data, which we included as a dummy variable. The statistical analysis was performed using SAS software version 9.4, R (R Core Team, 2020).
RESULTS
OS of Patients With Metastatic CRC Bearing Individual mutp53 GOF
OS associated with each individual GOF mutp53 of interest is shown in the Data Supplement (online only). Median OS was 24.3, 48.2, 14.5, 3.1, 26.4, and 23.8 months, respectively, for R175H, R248Q, R248W, R249S, R273H and R273L, and R282W. Median OS for the entire GOF subset was 24.3 months.
Comparison of Demographics and the Molecular Features Between mutp53 and wtp53 and Between mutp53 GOF and Non-GOF Patients
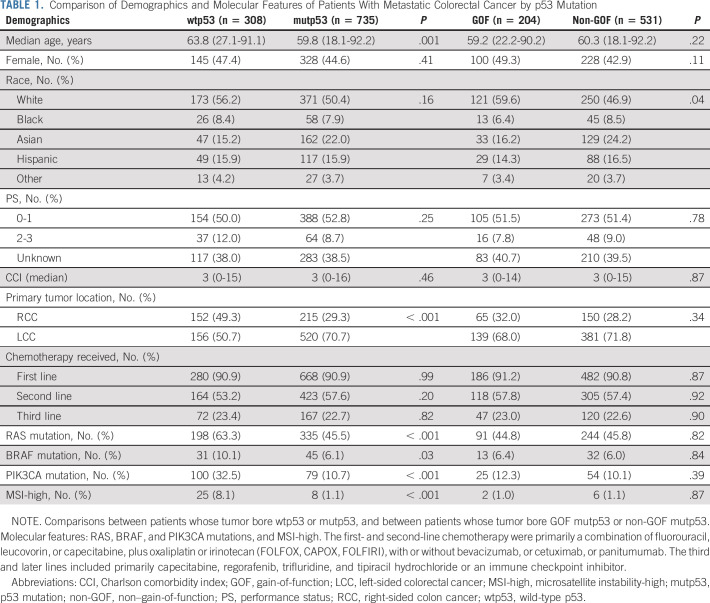
As depicted in Table 1, of 1,043 patients with metastatic CRC with StrataNGS performed, 308 had tumors with wtp53 and 735 had mutp53. The estimated median OS was 25.1 months for the entire cohort. Median age of wtp53 patients was higher than that of mutp53 patients, although the two groups were generally similar with respect to sex, ethnicity, PS, CCI, or chemotherapy received. A higher percent of wtp53 patients had RCC, and a higher percent had RAS (including both KRAS and NRAS), BRAF, and PIK3CA mutations, and MSI-high. Between the GOF and the non-GOF subsets, there was no statistically significant difference in median age, sex, PS, CCI, or chemotherapy received, but more GOF patients were White, whereas more non-GOF patients were Asian (P = .04). There was no significant difference in the percent of RAS, BRAF, and PIK3CA mutations, and MSI-high.
TABLE 1.
Comparison of Demographics and Molecular Features of Patients With Metastatic Colorectal Cancer by p53 Mutation
Comparison of Demographics and the Molecular Features Between LCC and RCC
As depicted in Table 2, compared with patients with LCC, patients with RCC were on average older, and a larger percent had PS of Eastern Cooperative Oncology Group 2-3, higher CCI, fewer patients received and have received second-line chemotherapy, and had more RAS, BRAF, and PIK3CA mutations, and MSI-high, even when separated by GOF, non-GOF, and wtp53. Patients with either BRAF or RAS mutation showed worse OS than patients with both BRAF and RAS wild-type (Data Supplement), with HR of 2.10 and 1.26, respectively, after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received (Data Supplement). OS of RCC was worse than OS of LCC (Data Supplement), with HR of 1.2 after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received (Fig 1).
TABLE 2.
Comparison of Demographics and Molecular Features of Patients With Metastatic LCC Versus RCC
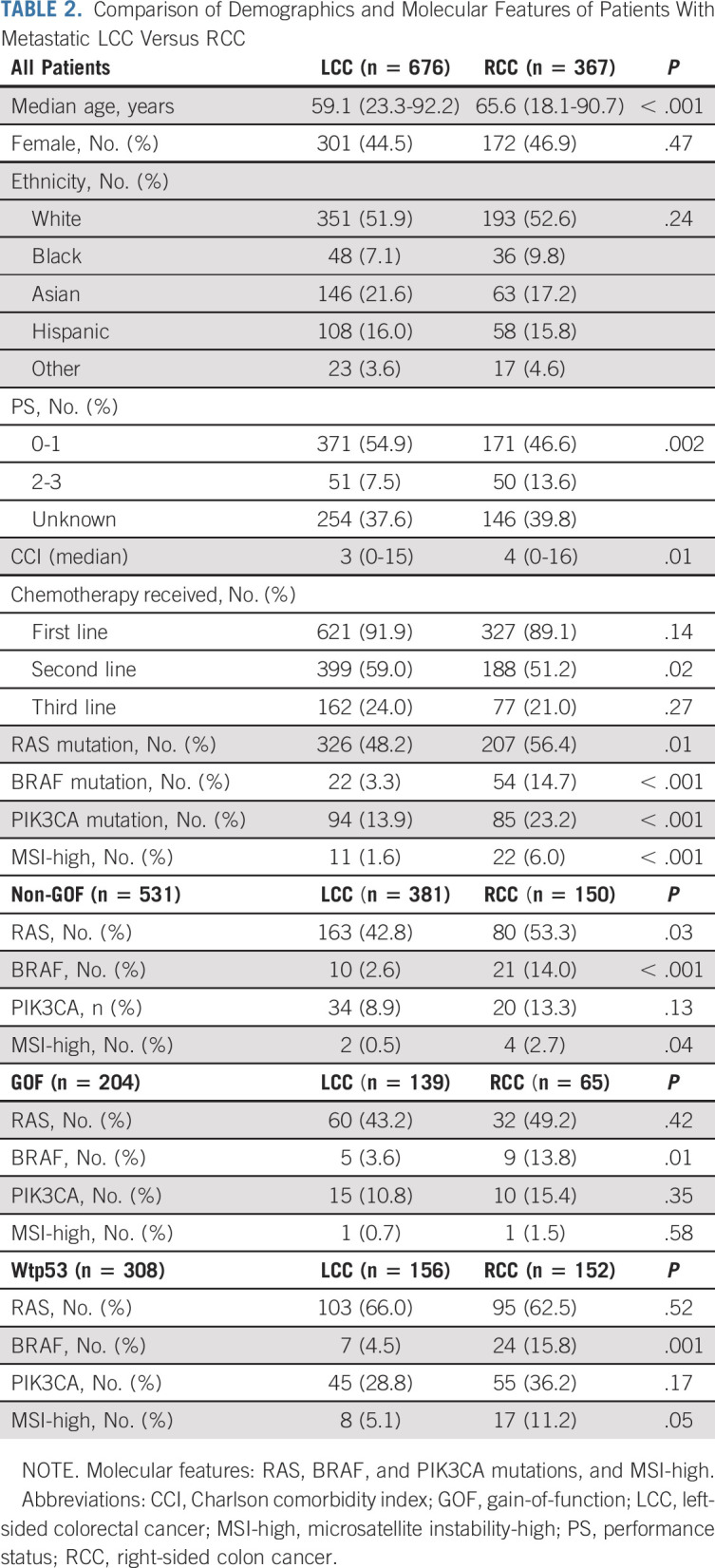
FIG 1.
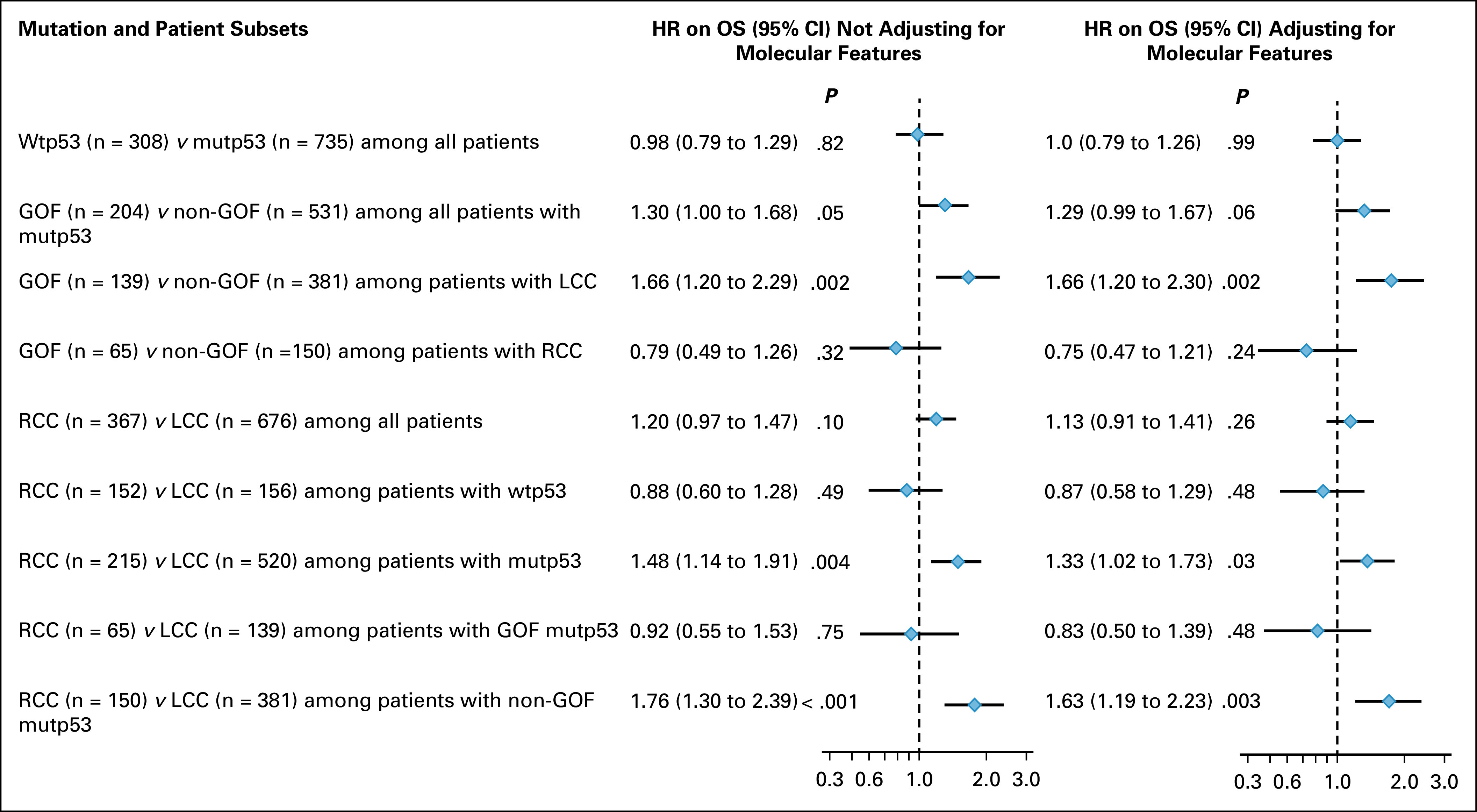
Forest plot for HR for OS associated with p53 mutation and sidedness. Multivariate analysis was performed using Cox regression models adjusting for age, sex, race, performance status, Charlson comorbidity index, chemotherapy received (left column), and further adjusting for molecular features including RAS, BRAF, and PIK3CA mutations, and MSI-high (right column). GOF, gain-of-function mutp53; HR, hazard ratio; LCC, left-sided colorectal cancer; MSI-high, microsatellite instability-high; mutp53, p53 mutation; non-GOF, non–gain-of-function mutp53; OS, overall survival; RCC, right-sided colon cancer; wtp53, wild-type p53.
Mutp53 GOF Was Associated With Worse Survival Than mutp53 Non-GOF Only in Patients With LCC But Not RCC
In the entire cohort, unadjusted and adjusted OS were similar between patients with wtp53 and patients with mutp53 (Data Supplement and Fig 1). Unadjusted OS curves showed little difference between patients with GOF and patients with non-GOF mutp53 (Data Supplement). However, after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received, OS of patients with GOF was approximately 30% worse than OS of patients with non-GOF (HR = 1.30 [95% CI, 1.00 to 1.68]), and this difference remained after further adjusting for the molecular features (RAS, BRAF, and PIK3CA mutations, and MSI-high; HR = 1.29 [95% CI, 0.99 to 1.67]; Fig 1). Intriguingly, OS of patients with LCC that bore mutp53 GOF was significantly worse than OS of patients with LCC that bore mutp53 non-GOF (median OS 24.6 v 33.5 months, P = .01; Fig 2A), and this difference remained after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received (P = .002; HR = 1.66 [95% CI, 1.20 to 2.29]) and after further adjusting for the molecular features (P = .002; HR = 1.66 [95% CI, 1.20 to 2.30]; Fig 1). By contrast, OS of patients with RCC that bore mutp53 GOF was not worse than OS of patients with RCC that bore mutp53 non-GOF (median OS 23.6 v 13.8 months, P = .08; Fig 2B), even after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received (HR = 0.79 [95% CI, 0.49 to 1.26]) and after further adjusting for the molecular features (HR = 0.73 [95% CI, 0.47 to 1.21]; Fig 1).
FIG 2.
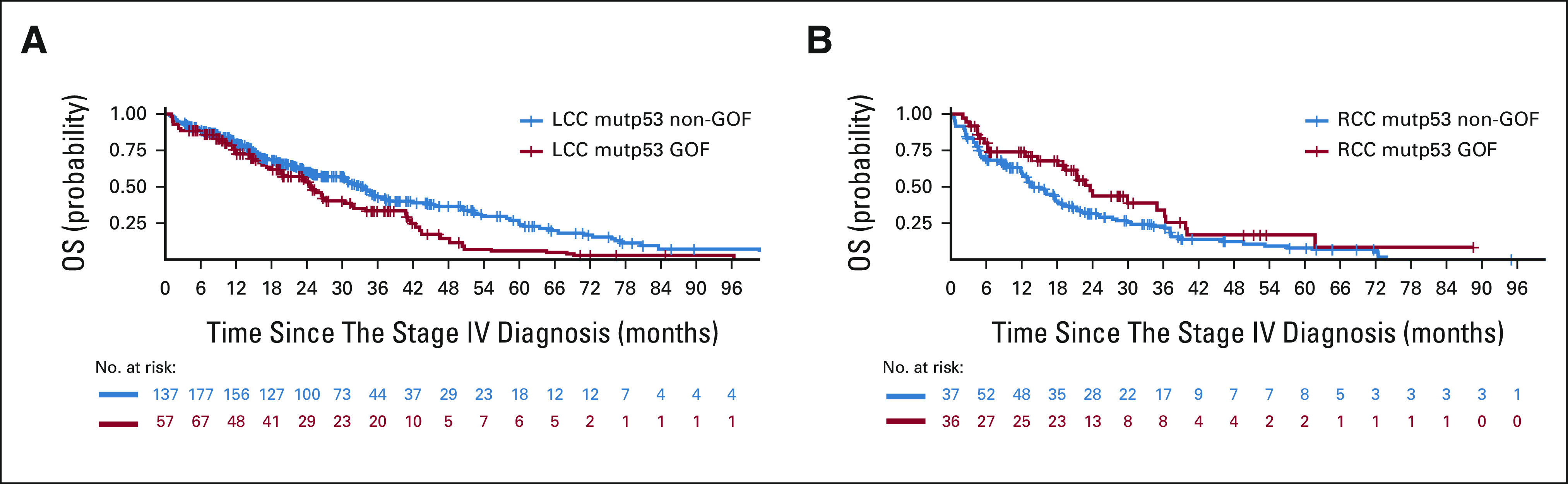
(A) Kaplan-Meier OS curves for patients with metastatic LCC that bore GOF versus non-GOF mutp53. Median OS was 24.6 (GOF, n = 139) versus 33.5 (non-GOF, n = 381) months, P = .01. (B) Kaplan-Meier OS curves for patients with metastatic RCC with GOF versus non-GOF p53 mutp53. Median OS was 23.6 (RCC mutp53 GOF, n = 65) versus 13.8 (RCC mutp53 non-GOF, n = 150) months, P = .08. GOF, gain-of-function; LCC, left-sided colorectal cancer; mutp53, p53 mutation; non-GOF, non–gain-of-function; OS, overall survival; RCC, right-sided colon cancer.
Mutp53 Non-GOF But Not GOF Was Associated With Worse Survival of Patients With Metastatic RCC Versus LCC
There was no meaningful OS difference between patients with RCC and patients with LCC that bore wtp53, either unadjusted or adjusted (Data Supplement and Fig 1). However, there was a significant OS difference between patients with RCC and patients with LCC that bore mutp53 (median OS 17.6 v 30.7 months, P < .001; Fig 3A), with HR of 1.48 (P = .004; 95% CI, 1.14 to 1.91) after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received, and HR of 1.33 (P = .03; 95% CI, 1.02 to 1.73) after further adjusting for the molecular features (Fig 1). Interestingly, there was no meaningful OS difference between patients with RCC and LCC whose CRC bore mutp53 GOF, either unadjusted (Fig 3B), or adjusted for age, sex, ethnicity, PS, CCI, and chemotherapy received or further adjusted for the molecular features (Fig 1). By contrast, there was a dramatic OS difference between patients with RCC and LCC whose CRC bore mutp53 non-GOF (median OS 13.8 v 33.5 months, P < .001; Fig 3C), and this OS difference remained highly significant after adjusting for age, sex, ethnicity, PS, CCI, and chemotherapy received (HR = 1.76 [95% CI, 1.30 to 2.39]) and after further adjusting for the molecular features (HR = 1.63 [95% CI, 1.19 to 2.23]; Fig 1).
FIG 3.
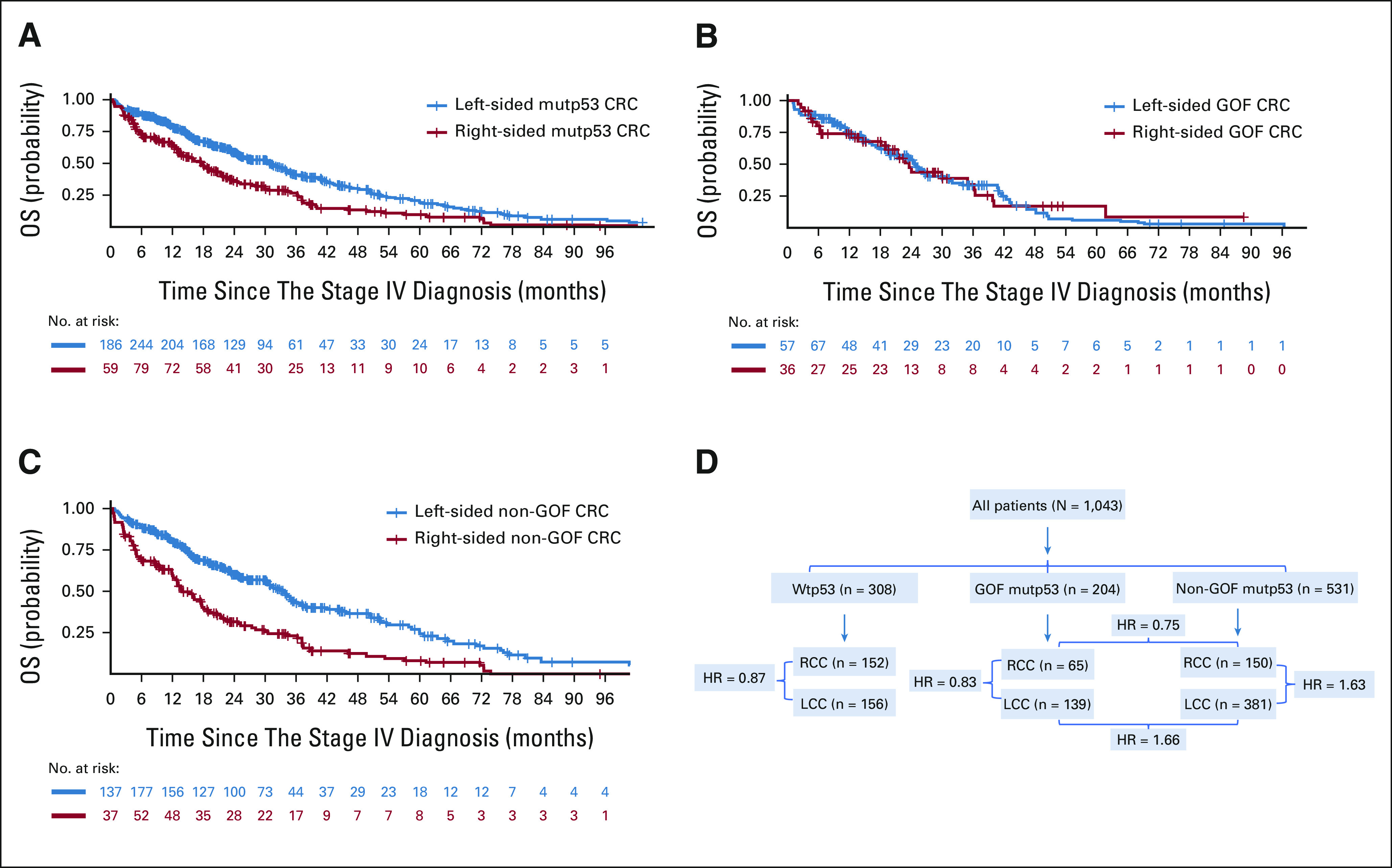
(A) Kaplan-Meier OS curves for patients with metastatic RCC versus LCC that bore mutp53. Median OS was 17.6 (right-sided mutp53 CRC, n = 215) versus 30.7 (left-sided mutp53 CRC, n = 520) months, P < .001. (B) Kaplan-Meier OS curves for patients with metastatic RCC versus LCC that bore gain-of-function p53 mutation (GOF). Median OS was 23.6 (right-sided GOF CRC, n = 65) versus 24.6 (left-sided GOF CRC, n = 139) months, P = .96. (C) Kaplan-Meier OS curves for patients with metastatic RCC versus LCC that bore non–gain-of-function p53 mutation (non-GOF). Median OS was 13.8 (right-sided non-GOF CRC, n = 150) versus 33.5 (left-sided non-GOF CRC, n = 381) months, P < .001. (D) Flow diagram illustrating key findings. HR = 0.83 for RCC versus LCC among patients with GOF mutp53, while 1.63 among patients with non-GOF mutp53; HR = 0.75 for GOF versus non-GOF among patients with RCC, while 1.66 among patients with LCC. CRC, colorectal cancer; GOF mutp53, gain-of-function p53 mutations; HR, hazard ratio; LCC, left-sided colorectal cancer; non-GOF mutp53, non–gain-of-function p53 mutations; OS, overall survival; RCC, right-sided colon cancer; Wtp53, p53 wild-type.
The comparisons between RCC versus LCC among patients with non-GOF mutp53, between GOF versus non-GOF among patients with LCC, and between RCC versus LCC among patients with mutp53 meet statistical significance on the basis of false discovery rate (Data Supplement).35 We have illustrated the key findings in the flow diagram (Fig 3D).
DISCUSSION
In this study in which p53 mutations were classified into GOF or non-GOF, we have uncovered unexpected associations between mutp53 and sidedness-dependent prognosis of patients with metastatic CRC: GOF versus non-GOF mutp53 was associated with inferior survival in patients with LCC but not RCC, whereas non-GOF but not GOF mutp53 was associated with poorer survival of RCC versus LCC. The magnitude of OS difference between patients with RAS or BRAF mutation versus RAS and BRAF wild-type, and between patients with RCC versus LCC was consistent with what has been reported in the literature,28,29,32,36,37 which supports the validity of our data set.
Our definition of GOF and non-GOF is likely incomplete as many of the individual mutp53 have not been studied for the GOF properties; however, such an approach allowed us to define the patients with metastatic CRC into distinct subsets with different prognosis. GOF mutp53 occur in high frequency and exhibit extensive loss of transcriptional function, whereas non-GOF mutp53 occur in low frequency, many of them preserve certain level of transcriptional function, and are capable of activating some of the p53-regulated genes.26,38
Most intriguingly, despite that patients with non-GOF mutp53 collectively had significantly better OS than patients with GOF mutp53, non-GOF mutp53 uniquely marked a population of patients with RCC, but not LCC, with poor survival. The poor survival of RCC appears to be a function of the non-GOF mutp53 patients, independent of GOF or wtp53. Non-GOF mutp53 is not likely to be directly responsible for the poor survival of patients with RCC, since patients with LCC had similar percent of non-GOF mutp53 and yet better survival than patients with RCC. It is more likely that non-GOF mutp53 is a biomarker for RCC and contributes to promoting the oncologic behavior by cooperating with other unidentified molecular lesions that play major roles in its poor survival. These molecular lesions could include components of the pathways targeted by high-fat diet, bile acids, CRC-associated microbiome, and other carcinogenic insults.39-41 In the non-GOF subset, the patients with RCC had significantly higher percent of RAS and BRAF mutations than the patients with LCC; however, this does not explain the worse OS of the non-GOF mutp53 RCC patients, because in the GOF mutp53 and the wtp53 subsets, the patients with RCC had similarly higher percent of BRAF mutation and yet similar OS compared with the patients with LCC. Importantly, even after further adjusting for RAS, BRAF, and PIK3CA mutations, and MSI-high, the OS of the RCC non-GOF patients remained dramatically worse than the OS of the LCC non-GOF patients.
Our study sheds new light for understanding the sidedness-dependent prognosis. Embryonically, right-sided colon arises from the mid-gut, whereas left-sided colon and the rectum from the hind-gut.42 How this embryonic patterning mechanism is related to the distinct role of mutp53 GOF and non-GOF in these two anatomically defined CRC subsets would be interesting to investigate. The p53 mutation pattern, either somatic or germline, exhibits tissue-specific preferences.38,43 There are differences in the gene expression profile between normal right colon epithelium and left colon epithelium; however, it is not known as to whether the intestinal stem cells of the right versus left colon possess distinct sensitivity to GOF and non-GOF mutp53.27 This could potentially be studied in mice by knocking-in GOF and non-GOF mutp53 separately into LGR5+ cells or using organoids in cultures.44 RCC is associated more frequently with BRAF mutation, MSI-high, CpG methylation, and resistance to anti-EGFR therapy, whereas LCC is associated more frequently with mutp53, ERBB2, and EGFR amplification, and response to anti-EGFR therapy.27 There is evidence that RCC but not LCC is associated with mucosal bacteria biofilms in the gut.45-48 Kadosh et al49 showed that gut microbiome could switch the action of GOF mutp53 R172H from being tumor-suppressive to oncogenic by activating WNT pathways, but only in the mouse jejunum and not ileum. Our findings may also be valuable in aiding the classification of molecular subtypes of CRC.27,50,51 In addition, our findings suggest that a reanalysis, using mutp53 GOF and non-GOF as additional biomarkers, of the previously completed large clinical trials evaluating the efficacy of cetuximab and bevacizumab plus chemotherapy may reveal additional insight.31,32,36,37,52-54 Future CRC clinical trials may benefit from incorporating mutp53 GOF and non-GOF as biomarkers in the design.
This study has several strengths. Our data set is relatively large with more than 1,000 patients who received comprehensive primary and specialty services from a large integrated health care system that consists of 21 medical centers. In addition, the diverse membership is relatively stable, and electronic records capture virtually all encounters, diagnoses, and procedures. Our study also has limitations. It is a retrospective study, and a number of patients did not have StrataNGS until several months to years after their diagnosis of metastatic disease. Nonetheless, we used appropriate statistical methods to address this issue.34 Also, the number of patients with some specific GOF mutp53 was small, limiting our ability to examine potential heterogeneity of OS findings across mutations. In addition, the follow-up length for some patients remained relatively short.
In summary, our study suggests that the functional classification of p53 mutations into GOF and non-GOF could help define the prognosis of metastatic CRC by sidedness and adds to the toolbox of the useful biomarkers. If confirmed, our findings could have important implications in clinical practice, future clinical trial design, and understanding the mechanism of CRC oncogenesis.
ACKNOWLEDGMENT
This paper is dedicated to the memory of Wumei and Daosheng Pan. The authors thank A. Boroian for support in manuscript preparation.
Stacey Alexeeff
Employment: The Permanente Medical Group
Wenwei Hu
Patents, Royalties, Other Intellectual Property: A patent application has been submitted (PCT/US20/20141). Title: LIF therapy for inducing intestinal epithelial cell regeneration
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part in two abstracts at the 2021 ASCO Virtual Annual Meeting, June 4-8, 2021.
SUPPORT
Supported by The Permanente Medical Group. M.P. was partly supported by the Jiayan Foundation. W.H. was supported by NIH/NCI R01CA260837.
DATA SHARING STATEMENT
Kaiser Permanente Northern California (KPNC) Institutional Review Board has not provided approval for StrataNGS data on individual patients used in this study to be placed in a public access repository. However, researchers can request access to use this study data by contacting the DOR Data Sharing Workgroup at DOR-DataSharingWorkgroup@kp.org.
AUTHOR CONTRIBUTIONS
Conception and design: Minggui Pan, Sachdev Thomas, Laurel A. Habel
Financial support: Minggui Pan
Administrative support: Minggui Pan, Pam Tse, Elaine Chung, Sachdev Thomas
Provision of study materials or patients: Minggui Pan, Chen Jiang, Ninah Achacoso, Thach-Giao Truong, Laurel A. Habel
Collection and assembly of data: Minggui Pan, Pam Tse, Ninah Achacoso, Aleyda V. Solorzano, Elaine Chung, Thach-Giao Truong, Amit Arora, Jennifer Marie Suga, Laurel A. Habel
Data analysis and interpretation: Minggui Pan, Chen Jiang, Ninah Achacoso, Stacey Alexeeff, Wenwei Hu, Amit Arora, Tilak Sundaresan, Sachdev Thomas, Laurel A. Habel
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
TP53 Gain-of-Function and Non–Gain-of-Function Mutations Are Differentially Associated With Sidedness-Dependent Prognosis in Metastatic Colorectal Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Stacey Alexeeff
Employment: The Permanente Medical Group
Wenwei Hu
Patents, Royalties, Other Intellectual Property: A patent application has been submitted (PCT/US20/20141). Title: LIF therapy for inducing intestinal epithelial cell regeneration
No other potential conflicts of interest were reported.
REFERENCES
- 1.Baker SJ, Vogelstein B: p53: a tumor suppressor hiding in plain sight. J Mol Cell Biol 11: 536–5382019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane D, Levine A: p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol 2: a000893.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller PA, Vousden KH: Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25: 304–3172014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine AJ: Surfing the p53 network. Nature 408: 307–3102000 [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network : Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–3372012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malkin D: Li-fraumeni syndrome. Genes Cancer 2: 475–4842011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols KE, Malkin D: Genotype versus phenotype: The Yin and Yang of germline TP53 mutations in Li-Fraumeni syndrome. J Clin Oncol 33: 2331–23332015 [DOI] [PubMed] [Google Scholar]
- 8.Kastenhuber ER, Lowe SW: Putting p53 in context. Cell 170: 1062–10782017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyi VA, Ak P, Markert E, et al. : The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2: a001198.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vousden KH, Prives C: Blinded by the light: The growing complexity of p53. Cell 137: 413–4312009 [DOI] [PubMed] [Google Scholar]
- 11.Levine AJ, Oren M: The first 30 years of p53: Growing ever more complex. Nat Rev Cancer 9: 749–7582009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Liu J, Xu D, et al. : Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol 12: 674–6872020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller PA, Vousden KH: p53 mutations in cancer. Nat Cell Biol 15: 2–82013 [DOI] [PubMed] [Google Scholar]
- 14.Brosh R, Rotter V: When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer 9: 701–7132009 [DOI] [PubMed] [Google Scholar]
- 15.Olivier M, Hollstein M, Hainaut P: TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baugh EH, Ke H, Levine AJ, et al. : Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 25: 154–1602018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmer D, Pati S, Zambetti G, et al. : Gain of function mutations in p53. Nat Genet 4: 42–461993 [DOI] [PubMed] [Google Scholar]
- 18.Lang GA, Iwakuma T, Suh YA, et al. : Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861–8722004 [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Qian J, Hu Y, et al. : Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep 4: 4223.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanel W, Marchenko N, Xu S, et al. : Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ 20: 898–9092013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz-Heddergott R, Stark N, Edmunds SJ, et al. : Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits Stat3-mediated tumor growth and invasion. Cancer Cell 34: 298–314.e2972018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue X, Wu F, Li Y, et al. : Gain of function mutant p53 protein activates AKT through the Rac1 signaling to promote tumorigenesis. Cell Cycle 19: 1338–13512020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue X, Zhang C, Zhao Y, et al. : Gain-of-function mutant p53 activates small GTPase Rac1 through SUMOylation to promote tumor progression. Genes Dev 31: 1641–16542017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooks T, Pateras IS, Tarcic O, et al. : Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 23: 634–6462013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Zhang C, Yue X, et al. : Pontin, a new mutant p53-binding protein, promotes gain-of-function of mutant p53. Cell Death Differ 22: 1824–18362015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oren M, Rotter V: Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2: a001107.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MS, Menter DG, Kopetz S: Right versus left colon cancer biology: Integrating the consensus molecular subtypes. J Natl Compr Canc Netw 15: 411–4192017 [DOI] [PubMed] [Google Scholar]
- 28.Venook AP: Right-sided vs left-sided colorectal cancer. Clin Adv Hematol Oncol 15: 22–242017 [PubMed] [Google Scholar]
- 29.Petrelli F, Tomasello G, Borgonovo K, et al. : Prognostic survival associated with left-sided vs right-sided colon cancer: A systematic review and meta-analysis. JAMA Oncol 3: 211–2192017 [DOI] [PubMed] [Google Scholar]
- 30.Loupakis F, Yang D, Yau L, et al. : Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 107: dju427.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venook AP, Niedzwiecki D, Lenz HJ, et al. : Effect of first-line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild-type advanced or metastatic colorectal cancer: A randomized clinical trial. JAMA 317: 2392–24012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tejpar S, Stintzing S, Ciardiello F, et al. : Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol 3: 194–2012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller TI, Zoumberos NA, Johnson B, et al. : A genomic survey of sarcomas on sun-exposed skin reveals distinctive candidate drivers and potentially targetable mutations. Hum Pathol 102: 60–692020 [DOI] [PubMed] [Google Scholar]
- 34.Hernan MA, Sauer BC, Hernandez-Diaz S, et al. : Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J Clin Epidemiol 79: 70–752016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Drai D, Elmer G, et al. : Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–2842001 [DOI] [PubMed] [Google Scholar]
- 36.Modest DP, Ricard I, Heinemann V, et al. : Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol 27: 1746–17532016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stintzing S, Miller-Phillips L, Modest DP, et al. : Impact of BRAF and RAS mutations on first-line efficacy of FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab: Analysis of the FIRE-3 (AIO KRK-0306) study. Eur J Cancer 79: 50–602017 [DOI] [PubMed] [Google Scholar]
- 38.Bouaoun L, Sonkin D, Ardin M, et al. : TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum Mutat 37: 865–8762016 [DOI] [PubMed] [Google Scholar]
- 39.Beyaz S, Mana MD, Roper J, et al. : High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531: 53–582016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu T, Coulter S, Yoshihara E, et al. : FXR regulates intestinal cancer stem cell proliferation. Cell 176: 1098–1112.e10182019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ternes D, Karta J, Tsenkova M, et al. : Microbiome in colorectal cancer: How to get from meta-omics to mechanism? Trends Microbiol 28: 401–4232020 [DOI] [PubMed] [Google Scholar]
- 42.Stintzing S, Tejpar S, Gibbs P, et al. : Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer 84: 69–802017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freed-Pastor WA, Prives C: Mutant p53: One name, many proteins. Genes Dev 26: 1268–12862012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gehart H, Clevers H: Tales from the crypt: New insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 16: 19–342019 [DOI] [PubMed] [Google Scholar]
- 45.Tomkovich S, Dejea CM, Winglee K, et al. : Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 129: 1699–17122019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dejea CM, Wick EC, Hechenbleikner EM, et al. : Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA 111: 18321–183262014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson CH, Dejea CM, Edler D, et al. : Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab 21: 891–8972015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomkovich S, Gharaibeh RZ, Dejea CM, et al. : Human colon mucosal biofilms and murine host communicate via altered mRNA and microRNA expression during cancer. mSystems 10.1128/mSystems.00451-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadosh E, Snir-Alkalay I, Venkatachalam A, et al. : The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 586: 133–1382020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guinney J, Dienstmann R, Wang X, et al. : The consensus molecular subtypes of colorectal cancer. Nat Med 21: 1350–13562015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dienstmann R, Vermeulen L, Guinney J, et al. : Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 17: 268.2017 [DOI] [PubMed] [Google Scholar]
- 52.Innocenti F, Ou FS, Qu X, et al. : Mutational analysis of patients with colorectal cancer in CALGB/SWOG 80405 identifies new roles of microsatellite instability and tumor mutational burden for patient outcome. J Clin Oncol 37: 1217–12272019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinemann V, von Weikersthal LF, Decker T, et al. : FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol 15: 1065–10752014 [DOI] [PubMed] [Google Scholar]
- 54.Stintzing S, Modest DP, Rossius L, et al. : FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): A post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol 17: 1426–14342016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Kaiser Permanente Northern California (KPNC) Institutional Review Board has not provided approval for StrataNGS data on individual patients used in this study to be placed in a public access repository. However, researchers can request access to use this study data by contacting the DOR Data Sharing Workgroup at DOR-DataSharingWorkgroup@kp.org.