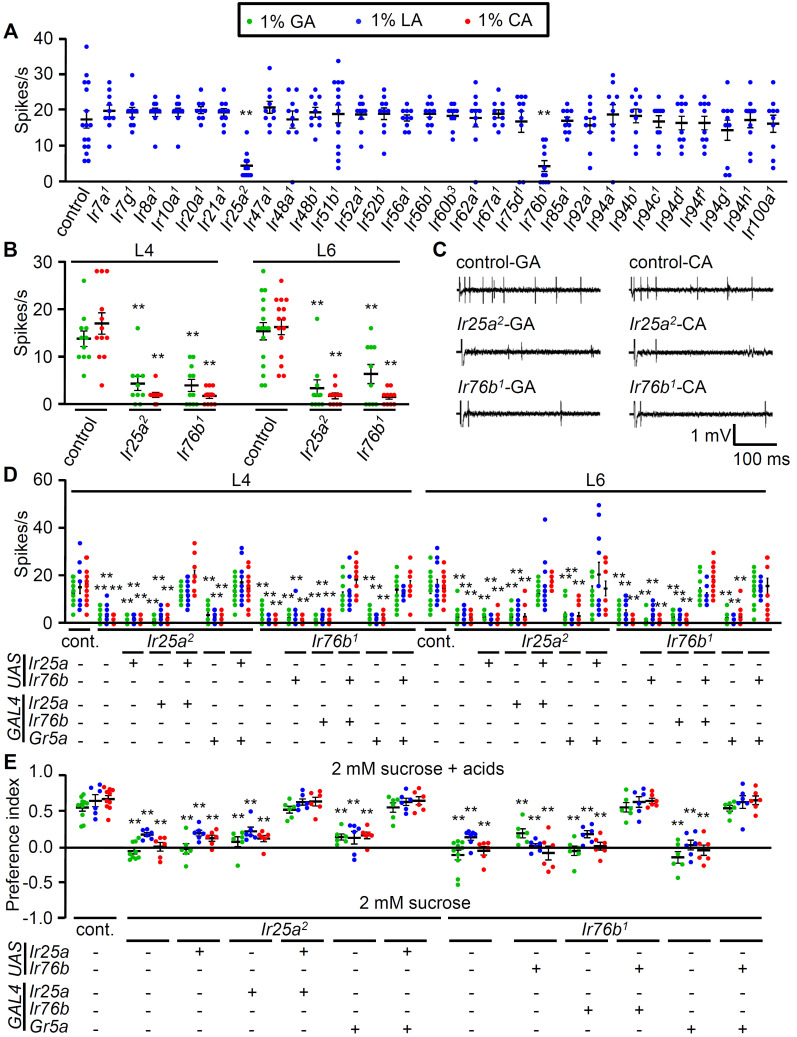
Fig. 3. IR25a and IR76b are required for sensing attractive carboxylic acids in sweet-sensing GRNs.
(A) Screening candidate Ir mutants from L6 sensilla stimulated with 1% LA (n = 10-14). (B) Neuronal firing responses of control, Ir25a2, and Ir76b1 from L4 and L6 sensilla stimulated with 1% GA and 1% CA (n = 10-16). (C) Representative sample traces of (A) and (B). (D) Electrophysiological rescue of Ir25a2 and Ir76b1 mutant defects elicited by the indicated acids from L4 and L6 sensilla using broadly expressed specific GAL4s (Ir25a-GAL4 and Ir76b-GAL4). Gr5a-GAL4 (sweet-sensing GRNs marker) was also used to drive both UAS lines. +/- indicate the presence or absence of the transgene, respectively (n = 10-14). cont., control. (E) Behavioral rescue of Ir25a2 and Ir76b1 mutant deficits to acid-attraction with the indicated acids in the binary food choice assay. UAS-Ir25a was driven by Ir25a-GAL4 or Gr5a-GAL4 in Ir25a2 mutant while UAS-Ir76b was driven by Ir76b-GAL4 or Gr5a-GAL4 in Ir76b1 mutant. +/- indicate the presence or absence of the transgene, respectively (n = 6-8). All error bars represent the SEM. Multiple sets of data were compared using single-factor ANOVA coupled with Scheffe’s post hoc test. Asterisks indicate statistical significance compared to the control (**P < 0.01).