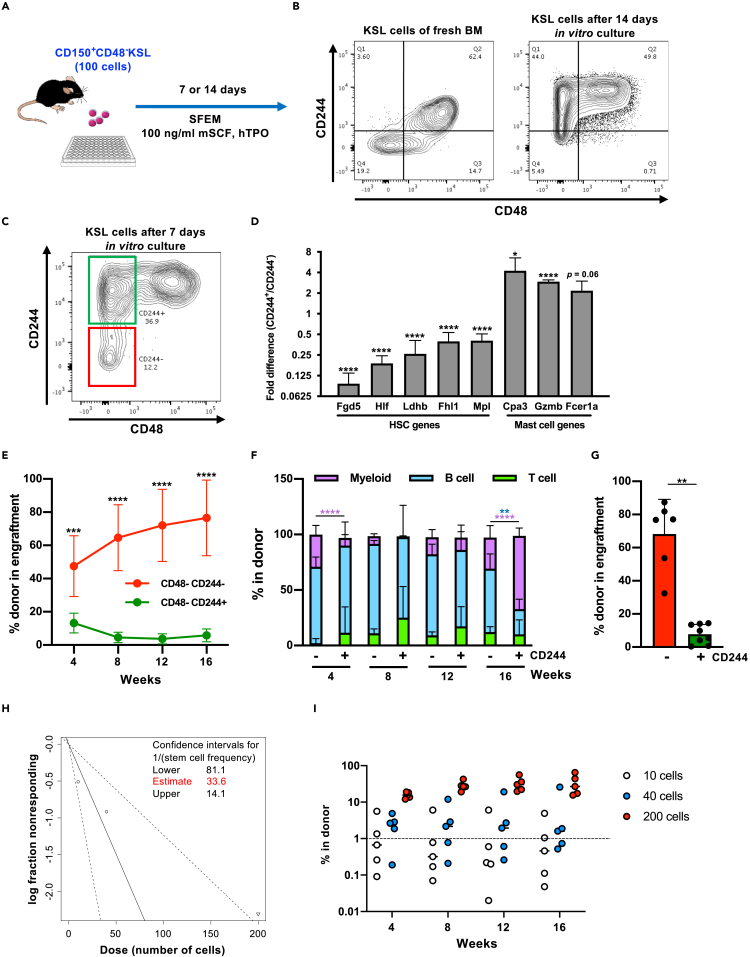
Figure 3.
CD244 expression divides CD48−KSL cells into functionally distinct subpopulations after in vitro culture
(A) Experimental design of the in vitro culture experiment. One hundred CD48−KSL cells were sorted from BM of young mice and cultured in Stemspan SFEM medium supplemented with 100 ng/mL mSCF and 100 ng/mL hTPO for 7 days.
(B) Expression patterns of CD244 and CD48 on the cell surface of fresh and 14 days cultured KSL cells. Representative FACS plots on KSL population are shown.
(C and D) qRT-PCR analysis for HSC-related genes and mast cell-related genes in CD244+CD48−KSL cells compared with the CD244+CD48−KSL counterpart. Representative FACS plot and gating of CD244+CD48−KSL cells (green) and CD244−CD48−KSL cells (red) on 7 days cultured KSL cells are shown in (C). Relative expression levels of genes in CD244+CD48−KSL cells to CD244−CD48−KSL are shown in (D).
(E) Competitive reconstitution assay. After 7 days’ culture, two subpopulations were sorted and 1,000 of CD244- or 1,500 of CD244+CD48−KSL cells were separately transplanted into lethally irradiated recipient mice (seven mice) with 2 × 105 total BM cells (competitor). Chimerism was monitored by analyzing PB every month. Significance was calculated using Student's t test at each time point. Mean ± SD from two independent experiments (n = 7) are displayed. ∗∗∗∗p < 0.0001.
(F) Lineage balance of donor-derived cells in the PB of recipient mice. Mean ± SD from two independent experiments (n = 6) are displayed. ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Each color represents different lineages.
(G) Analysis of BM from engrafted mice after 16 weeks. Chimerism in each cell fraction is shown. Significance was calculated using Student's t test. Mean ± SD from two independent experiments (n = 6) are displayed. ∗∗p < 0.01.
(H and I) Limiting dilution assay for CD244−CD48−KSL cells. CD244−CD48−KSL cells were re-sorted after 7 days’ in vitro culture and 200, 40, or 10 cells were transplanted to recipient mice in a competitive manner. Chimerism above 1% was judged as successful engraftment. The frequency of functional HSC was calculated using ELDA (http://bioinf.wehi.edu.au/software/elda/). Chimerism of individual mice is shown in (I).