Figure 1.
HIF signaling and glycolysis are unique characteristics in male EC on a proteomic level
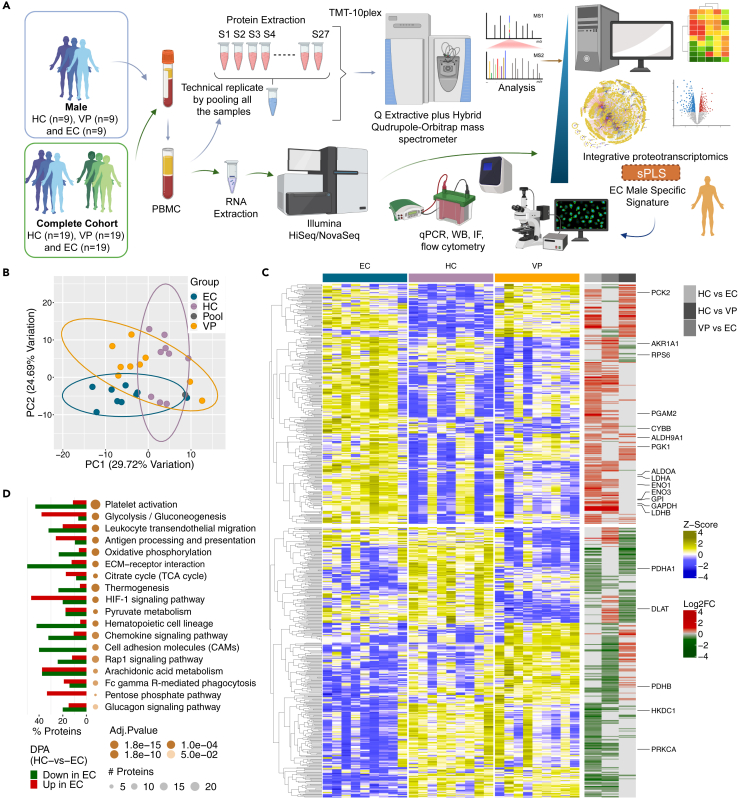
Study design and proteomic analysis.
(A) Study design and workflow of sample processing, data generation, and integrative analysis of proteomic and transcriptomic data. PBMCs were prepared from a cohort of male HIV-1-negative individuals (HC, n = 9), elite controllers (EC, n = 9), and viremic progressors (VP, n = 9); and protein lysates were analyzed on Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer while RNA was sent for Illumina HiSeq/NovaSeq sequencing together with a larger cohort of both male and female HC (n = 19), EC (n = 19), and VP (n = 19). Raw data processing, analysis, and data integration were performed by an in-house R script and sparse partial least squares (sPLS) regression and classification.
(B) Principal component analysis showing distribution of all the samples with respect to proteomics data. The data is plotted on 2D space after normalization and batch correction. The first two principal components capturing maximum of variances were used. Data ellipses were drawn at the level of 0.95.
(C) Heatmap visualizing quantile normalized and Z-scaled expression patterns of proteins significantly dysregulated in any of the pairwise comparisons among the study cohorts. Column annotations represent the cohorts and the different analysis pairs used for comparison. Proteins are clustered hierarchically based on the Euclidean distance.
(D) Visualization of functional analysis of significantly dysregulated proteins in EC compared to HC. The size of the bubbles is relative to the number of features within each pathway while the color gradients represent adjusted p value of the enrichment test. The bar graph denotes number of proteins increased or decreased in each pathway. See also Figure S1.