Figure 1.
Cell populations in the ovarian follicles of wild-type and obese mice
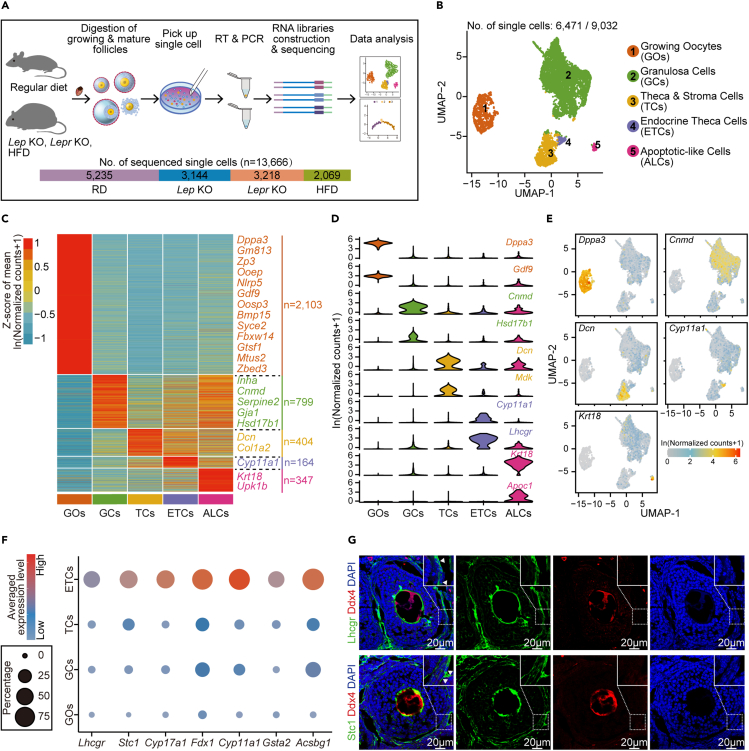
(A) Schematic representation of the mouse ovarian follicle single-cell RNA-seq workflow and the downstream analysis (top). The number of sequenced single cells for RD (regular diet), Lep KO, Lepr KO and HFD (high-fat diet) mice are shown as stacked bars (bottom).
(B) UMAP (Uniform Manifold Approximation and Projection) embedding visualization for 6,471 single cells from mouse ovarian follicles showing five major cell populations, including growing oocytes (GOs), granulosa cells (GCs), theca/stroma cells (TCs), endocrine theca cells (ETCs) and apoptotic-like cells (ALCs).
(C) Heatmap depicting DEGs among all identified cell populations derived from natural logarithm-scaled normalized counts based on the single-cell RNA-seq data. Gene expression levels were averaged and scaled.
(D) Violin plots showing expression levels of representative genes that were explicitly expressed in the indicated cell population.
(E) UMAP embedding visualization of cells in (B). Cells are color coded by expression levels of Dppa3, Cnmd, Dcn, Cyp11a1 or Krt18, respectively.
(F) Dot plot depicting the average expression levels of Lhcgr, Stc1, Cyp17a1, Fdx1, Cyp11a1, Gsta2, Acsbg1 in ETCs, TCs, GCs, and GOs, respectively. Averaged expression levels were derived from natural logarithm-scaled normalized counts based on the single-cell RNA-seq data. The percentage of cells expressing each gene is indicated by the size of the dot, and only cells with an expression level of greater than 1 for the indicated gene were counted.
(G) Immunofluorescence staining for Lhcgr or Stc1 in ovarian tissues from RD mice. Tissues were counterstained with Ddx4 and DAPI.
See also Table S1.