Figure 2.
Alternations in subgroup composition of ovarian GCs from the obese mice
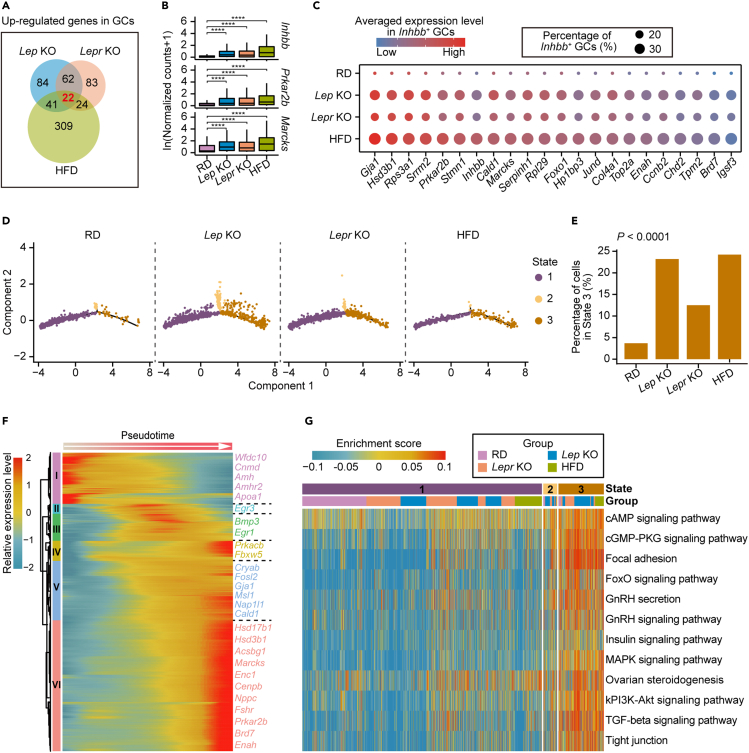
(A) Venn diagram showing the up-regulated genes in GCs overlapping between Lep KO, Lepr KO, and HFD mice compared with RD mice.
(B) Boxplots showing expression levels of Inhbb, Prkar2b and Marcks in GCs from RD, Lep KO, Lepr KO and HFD mice, respectively. The asterisks above indicate statistical significance. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (Wilcoxon rank-sum test).
(C) Dot plot depicting the average expression levels of 22 up-regulated genes identified in Figure 2A in Inhbb+ GCs.
(D) Pseudotime trajectory of granulosa cells in Figure 1B. Trajectory was inferred by Monocle 2. Cells are color coded by State inferred in the pseudotime analysis.
(E) Bar plot showing the percentage of GCs in State 3 from RD and the obese mice, respectively. The Chi-squared test was performed.
(F) Heatmap depicting genes with pseudotemporal expression patterns in GCs from RD and the obese mice, which were identified using Monocle2. Gene expression levels were scaled using Monocle2.
(G) Heatmap depicting representative pathways enriched in GCs from different states based on the results of GSVA (gene set variation analysis). Enrichment scores are scaled and color coded. Group and State for GCs are as indicated.