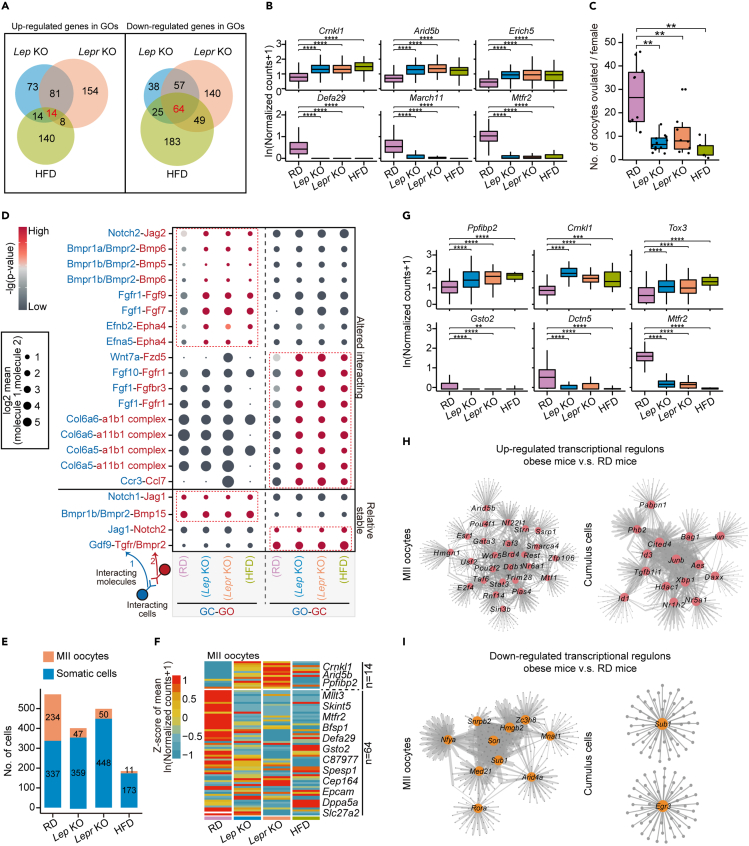
Figure 5.
Interactions between oocytes and granulosa cells in RD and obese mice
(A) Venn diagram showing the up-regulated (left) and down-regulated (right) genes in growing oocytes overlapping between Lep KO, Lepr KO, and HFD mice compared with RD mice.
(B) Boxplots showing expression levels of representative up-regulated genes (Crnkl1, Arid5b, Erich5) and down-regulated genes (Defa29, March11, Mtfr2) in growing oocytes in the obese mice compared with RD mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(C) Boxplot showing the number of ovulated oocytes in Lep KO (n = 12), Lepr KO (n = 10), and HFD (n = 5) mice is compromised compared with that of RD mice (n = 8). ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(D) Dot plot depicting representative ligand-receptor interactions between GOs and GCs. Interaction strength was measured using log2-scaled means of the average expression level of the ligand in the indicated cell type and receptor in the other cell type. The p values (permutation test) are color coded.
(E) Stacked bar plot showing the number of sequenced single cells in RD, Lep KO, Lepr KO, and HFD mice, grouped by MII oocytes and somatic cells.
(F) Heatmap showing the gene expression patterns in MII oocytes for 78 DEGs identified in Figure 5A.
(G) Boxplots showing expression levels of representative up-regulated genes (Ppfibp2, Crnkl1, Tox3) and down-regulated genes (Gsto2, Dctn5, Mtfr2) in MII oocytes in the obese mice compared with RD mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(H and I) Regulatory network of up- (H) and down-regulated (I) regulons in MII oocytes and cumulus cells in obese mice compared with RD mice. Regulons overlapping with DEGs identified in single-cell RNA-seq data (obese mice versus RD mice) are highlighted. The line size indicates the weight of the target regulon.
See also Table S5.