Abstract
Plasmablastic lymphoma (PBL) is a rare and clinically aggressive neoplasm that typically occurs in immunocompromised individuals, including those infected with human immunodeficiency virus (HIV) and solid organ allograft recipients. Most prior studies have focused on delineating the clinico-pathological features and genetic attributes of HIVrelated PBL, in which MYC deregulation, Epstein-Barr virus (EBV) infection and, more recently, mutations in JAK/STAT, MAP kinase, and NOTCH pathway genes have been implicated in disease pathogenesis. The phenotypic spectrum of post-transplant (PT)-PBL is not well characterized and data on underlying genetic alterations are limited. This led us to perform comprehensive histopathological and immunophenotypic evaluation and targeted sequencing of 18 samples from 11 patients (8 males, 3 females; age range, 12-76 years) with PT-PBL; eight de novo and three preceded by other types of post-transplant lymphoproliferative disorders. Post-transplant PBL displayed morphological and immunophenotypic heterogeneity and some features overlapped those of plasmablastic myeloma. Six (55%) cases were EBV positive and five (45%) showed MYC rearrangement by fluorescence in situ hybridization. Recurrent mutations in epigenetic regulators (KMT2/MLL family, TET2) and DNA damage repair and response (TP53, mismatch repair genes, FANCA, ATRX), MAP kinase (KRAS, NRAS, HRAS, BRAF), JAK/STAT (STAT3, STAT6, SOCS1), NOTCH (NOTCH1, NOTCH3, SPEN), and immune surveillance (FAS, CD58) pathway genes were observed, with the mutational profiles of EBV+ and EBV– cases exhibiting both similarities and differences. Clinical outcomes also varied, with survival ranging from 0-15.9 years after diagnosis. Besides uncovering the biological heterogeneity of PT-PBL, our study highlights similarities and distinctions between PT-PBL and PBL occurring in other settings and reveals potentially targetable oncogenic pathways in subsets of the disease.
Introduction
Plasmablastic lymphoma (PBL) is an uncommon and aggressive B-cell non- Hodgkin lymphoma characterized by a proliferation of cells with immunoblastic or plasmablastic morphology, occasionally with a component of mature plasma cells, and an immunophenotype indicative of terminal B-cell differentiation.1 PBL usually occurs in the context of immune dysregulation, which may be due to human immunodeficiency virus (HIV) infection, iatrogenic (e.g., after organ transplantation), congenital, or age-related (immune senescence).2 Prior studies of mostly HIV-assocated PBL have highlighted the prognostic significance of disease stage and Epstein-Barr virus (EBV) status2-4 and genomic analyses have revealed frequent MYC rearrangements, heterogeneous chromosome/DNA copy number abnormalities,5-7 variable transcriptional and microRNA profiles2,8-10 and, recently, recurrent mutations in JAK/STAT, MAPK and NOTCH pathway genes.7,11
PBL occurring after solid organ transplantation (PT-PBL) is an uncommon type of post-transplant lymphoproliferative disorder (PTLD), accounting for 6-7% of PTLD and constituting a minor fraction (5-14%) of all PBL.2-4,12,13 Only limited data regarding the pathological and molecular features of PT-PBL have been reported.2-4,12,14
In order to clarify the pathogenetic bases of PT-PBL, we performed morphological, immunophenotypic and molecular analyses, including targeted genomic sequencing, of a series of PT-PBL, comprising de novo PBL, both primary and recurrent tumors, and those preceded by other types of PTLD.
Methods
Case selection
We searched our departmental database for cases of PTLD diagnosed over the past 18 years (2002-2019) to select those fulfilling morphological and immunophenotypic features of PBL according to the current World Health Organization classification.1 Other types of PTLD preceding PT-PBL were also identified. Clinical and laboratory data were retrieved from electronic health records. This study was performed according to the principles of the Declaration of Helsinki and a protocol approved by the Institutional Review Board of Columbia University.
Morphology and immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) tissue sections were stained with hematoxylin and eosin for cytomorphological evaluation and semi-quantitative assessment of the percentage of mature plasma cells. Immunohistochemistry/in situ hybridization was performed to analyze expression of B- and plasma-cell antigens, the cellular microenvironment, a variety of biomarkers and the EBV status/latency profiles (see Online Supplementary Methods).
Immunoglobulin heavy chain (IGH) gene rearrangement analysis
Polymerase chain reaction analysis for immunoglobulin heavy chain (IGH) gene rearrangement was performed on DNA extracted from fresh or FFPE tissue using the BIOMED-2 primers, as described previously.15
Cytogenetic analysis
G-band karyotyping was performed on metaphase preparations obtained after unstimulated overnight culture. Fluorescence in situ hybridization (FISH) was performed on metaphase spreads or FFPE sections using TP53/CEP 17 and MYC/IGH/CEP8 probes (Abbott Molecular, Des Plaines, IL, USA) using standard methods. Two hundred cells per hybridization were evaluated. For interphase FISH analysis, the cut-off was 1% for IGH/MYC and 4% for TP53/CEP 17 alterations.
Targeted genomic sequencing
DNA was extracted from tumors and matched non-tumor tissue for sequencing a panel of 465 cancer-associated genes, as described previously16 (Online Supplementary Methods). Microsatellite instability (MSI) was also analyzed (Online Supplementary Methods).
Results
Clinical characteristics
We analyzed 18 samples from 11 patients (8 males, 3 females; median age 61 years; range, 12-76 years) with PTPBL, accounting for 11/177 (6%) of ‘destructive’B-cell PTLD and 11/98 (11%) of monomorphic B-cell PTLD diagnosed at our institution during the study period. PT-PBL occurred in recipients of heart (4/11, 36%), kidney (3/11, 27%), lung (3/11, 27%) and combined liver/kidney (1/11, 9%) allografts at a median of 9.6 years after transplantation (range, 0.6-11.9 years). The intestines were the most common sites of disease (6/11, 55%). Three patients had recurrent PBL and in three patients the PBL was preceded by another type of PTLD. Staging marrow biopsies, performed in seven patients, including three of five with EBV– PBL, showed no evidence of PBL/PTLD. Therapy and outcome data are summarized in Table 1 and details are provided in the Online Supplementary Data. Serum protein electrophoresis revealed low-level monoclonal paraproteins in four of seven (57%) patients with available results; none had lytic bone lesions on imaging. Results of pertinent laboratory tests and imaging studies are listed in Online Supplementary Table S2.
Morphologic and immunophenotypic features
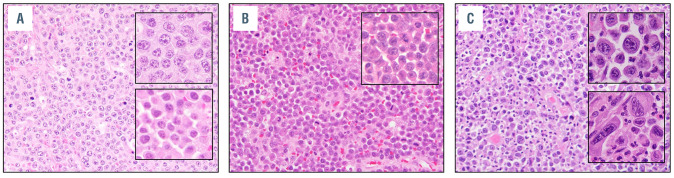
All PT-PBL showed diffuse infiltrates of large immunoblastic or plasmablastic cells (Figure 1). A minor component of small, more mature plasma cells (plasmacytic differentiation), comprising 10-20% of the neoplastic infiltrate, was seen in five (45%) cases (Figure 1A, Table 2). Some PBL had numerous tingible body macrophages, imparting a ‘starry sky’ appearance (Figure 1B) or multinucleated/ anaplastic cells (Figure 1C). Foci of necrosis were observed in six of 11 (55%) cases.
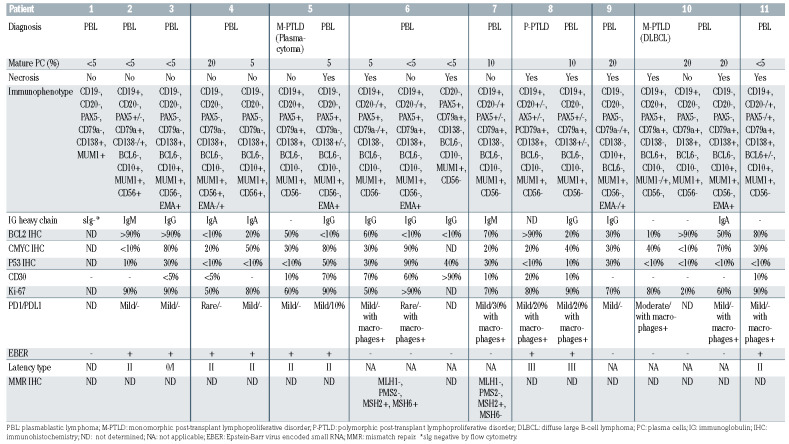
Details of the immunophenotypes of all cases are listed in Table 2 and flow cytometry results in Online Supplementary Table S3. Representative cases are illustrated in Figure 2. All PBL expressed MUM1/IRF4, nine of 11 (82%) were CD138+ and subsets showed B-cell antigen, CD10, CD56, PD-1 or PD-L1 expression. IgG, IgA or IgM was expressed by five (45%), two (18%), and two (18%) of the 11 cases. All evaluable PBL were positive for EMA and negative for Cyclin D1, CD117, HHV8 and ALK. Variable CD30 positivity was noted in seven of the 11 (64%) cases. The Ki-67 proliferation index ranged from 20 to >90% (median 90%). MYC expression ranged from <10% to 90% (median 45%), with six of ten (60%) cases showing ≥40% MYC expression. Two of the latter cases expressed BCL2 in ≥50% of cells (‘double expressors’). P53 overexpression was observed in five of ten (50%) cases. The immunoprofiles and/or proportions of cells expressing certain antigens differed in some PBL on recurrence. A mild to moderate infiltrate of reactive PD-1+ lymphocytes was observed in all ten cases evaluated and five of ten (50%) had PD-L1+ macrophages admixed.
Three PBL were preceded by other forms of PTLD; nodal EBV+ monomorphic PTLD (plasmacytoma) (case 5), duodenal EBV+ P-PTLD (case 8), and intestinal EBV– monomorphic PTLD (diffuse large B-cell lymphoma, DLBCL) (case 10).
Epstein-Barr virus status and latency profiles
The neoplastic cells were positive for EBER in six of 11 (55%) PBL, with four of these six (67%) showing latency II and one case each displaying latency 0/I and latency III profiles. Four EBV+ PBL and two EBV– PBL occurred in patients seropositive for EBV at the time of transplantation. EBV viremia at diagnosis was observed in all four EBV+ and two of five patients with EBV– PBL with available results (Online Supplementary Table S2).
Table 1.
Clinical features of post-transplant plasmablastic lymphomas.
IGH gene rearrangement analysis
All PT-PBL and both preceding monomorphic PTLD showed clonal IGH gene rearrangements. Clonal relatedness was established in all three recurrent PBL and between the PBL and prior monomorphic PTLD. The polymorphic PTLD demonstrated oligoclonal products.
Cytogenetic abnormalities
Cytogenetic findings are listed in Table 3. All three PBL with informative results showed complex karyotypes. IGHMYC rearrangements were detected in five of 11 (45%) cases (3 at diagnosis, 2 at recurrence). Multiple copies of MYC due to polyploidy were detected in two cases (concurrent with MYC rearrangement in 1 case). Three of six cases with MYC abnormalities (2 with rearrangements, 1 with gain) showed ≥40% MYC positivity by immunohistochemistry. PBL of patients who died of PTLD, and those who did not, revealed MYC abnormalities in two of four (50%) versus four of seven (57%) cases, respectively (P=1.0). Two cases demonstrated IGH rearrangements with unknown partners. Chromosome 17/TP53 abnormalities were detected in six; subclonal (30-40% of cells) monosomy 17 (1 EBV+, 1 EBV–), multiple copies (polyploidy) of chromosome 17 (2 EBV–), and TP53 deletion (1 EBV+, 1 EBV–). Loss of TP53 was detected by targeted genomic sequencing in two cases, including one case with insufficient material for FISH analysis.
No MYC or TP53 alterations were observed in the polymorphic PTLD or monomorphic PTLD (DLBCL) preceding PBL. Insufficient material precluded analysis of the PT-plasmacytoma.
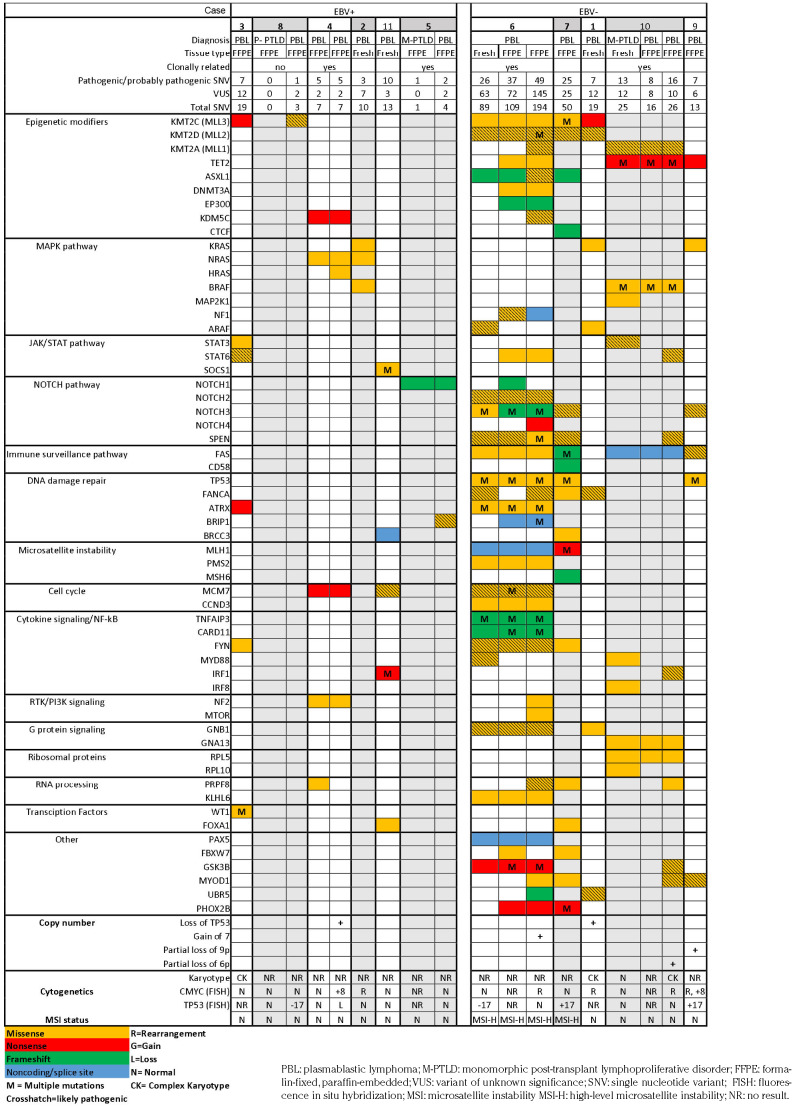
Targeted genomic sequencing
Pathogenic and likely pathogenic somatic non-synonymous single nucleotide variants (SNV) are listed in Table 3 and all, including variants of unknown significance (VUS), are listed in Online Supplementary Table S1. Excluding samples from two patients with high-level MSI, which exhibited up to 194 total SNV, the PBL harbored one to 16 pathogenic/ likely pathogenic (median 7) and three to 26 total SNV (median 13) including VUS. MSI status was confirmed by a polymerase chain reaction-based method. All four PBL samples classified as having high-level MSI (from patients 6 and 7) demonstrated loss of expression of at least two mismatch repair (MMR) proteins (Table 2). After factoring out cases with high level MSI, the number of variants was still higher in EBV– than EBV+ PBL, but the difference was not statistically significant (4.7 mean pathogenic/likely pathogenic and 9.3 total SNV in EBV+ vs. 10.0 mean pathogenic/likely pathogenic and 19.3 total SNV in EBV– cases; P=0.15 and P=0.09, respectively). There was no a statistically significant difference in the number of variants between patients who died of PBL and those who did not (4.7 mean pathogenic/likely pathogenic and 10.0 total SNV vs. 7.3 mean pathogenic/likely pathogenic and 14.0 total SNV; P=0.51 and P=0.6, respectively).
In our series, mutations were most frequent in epigenetic modifier genes, occurring in eight of 11 (73%) patients, and included recurrent mutations in the KMT2/MLL family of methyltransferases (KMT2C, n=5; KMT2D, n=3; and KMT2A, n=2), TET2 (n=3), ASXL1 (n=2), and KDM5C (n=2).
Figure 1.
Morphologic spectrum of post-transplant plasmablastic lymphomas. Representative hematoxylin and eosin (H&E)-stained sections of post-transplant plasmablastic lymphomas - from (A) case 4, showing a monotonous infiltrate of plasmablasts, with insets highlighting areas of plasmablastic morphology (top) and focal areas of plasmacytic differentiation (bottom), (B) case 5, showing numerous tingible body macrophages that impart a “starry sky” appearance, and (C) case 7, showing pleomorphic morphology, with insets displaying areas of plasmablastic morphology (top) and anaplastic-appearing multinucleated cells (bottom).
DNA damage response and repair pathway genes were mutated in seven of 11 (64%) cases, including four of five (80%) EBV– and three of six (50%) EBV+ PBL. Three of seven (43%) cases, all EBV–, including both casese with high-level MSI, showed multiple TP53 mutations. One case with concomitant subclonal monosomy 17 showed 30% P53+ cells, as did two with concurrent chromosome 17 gains, suggesting bi/multiallelic inactivation (Tables 2 and 3). Chromosome 17/TP53 abnormalities were more frequent in PBL exhibiting plasmacytic differentiation (4/5, 80%) than in those lacking it (2/6, 33%), although the difference was not statistically significant (P=0.24). PBL with high-level MSI (cases 6 and 7) had mutations in the MMR genes: MLH1 and PMS2 or MSH6, respectively. Immunohistochemistry showed that these cases also had loss of MMR proteins (Table 2) as well as a high mutational burden. Other recurrently- mutated genes included BRCC3, ATRX, FANCA, and BRIP1.
Mutations in the mitogen-activated protein kinase (MAPK) pathway genes were observed in six of 11 (55%) cases. Eight of nine mutations, occurring in KRAS, NRAS, HRAS, and BRAF, are well-known activating hotspot mutations (RAS codons 12, 13, and 61, and BRAF codons 469 and 601). Four cases harbored mutations in multiple genes, including one case with concomitant KRAS G13D, NRAS G12D, and BRAF G469V mutations.
Mutations in NOTCH signaling pathway genes were detected in five of 11 (45%) cases. Putative gain- or loss-offunction mutations in genes encoding the NOTCH family of proteins were the most frequent. SPEN, a negative regulator of NOTCH signaling, was mutated in three cases. Many of the NOTCH pathway mutations were in PBL with MMR defects and one case exhibited mutations in multiple NOTCH pathway members at different time points.
Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway genes, including STAT6 (n=3), STAT3 (n=2) and SOCS1 (n=1), were mutated in four of 11 (36%) cases.
Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway genes, including STAT6 (n=3), STAT3 (n=2) and SOCS1 (n=1), were mutated in four of 11 (36%) cases.
The monomorphic PTLD (plasmacytoma), which harbored a NOTCH1 mutation, acquired a BRIP1 mutation upon transformation. In contrast, some of the mutations observed in the monomorphic PTLD (DLBCL), including a STAT3 variant, were not detected in the clonally related transformed PBL, suggesting divergent evolution from a common ancestor. No variants were identified in the polymorphic PTLD preceding one PBL. The recurrent PBL showed both acquisition and loss of variants; the case with recurrent high-level MSI PBL (case 6) showed an increasing mutational burden over time.
Discussion
Our understanding of PT-PBL pathogenesis is limited and its molecular underpinnings are largely inferred from those reported for HIV-related PBL. Transcriptional analyses have revealed upregulation of JAK-STAT pathway genes and similarities between PBL and plasma cell neoplasms (multiple myeloma [MM] and extra-osseous plasmacytoma) and differences between immunocompetent DLBCL and PBL, the latter displaying increased expression of MYC and MYB target genes and genes regulating plasma cell differentiation and decreased expression of B-cell receptor and NF-kB signaling pathway genes.7,9 However, microRNA expression analysis has suggested two different subclasses of PBL, resembling either Burkitt lymphoma or extra-osseous plasmacytoma. 10 Chang et al. documented overlapping chromosomal aberrations between PBL and immunocompetent DLBCL as well as PBL-specific segmental gains at chromosomes 1p and 1q by array comparative genomic hybridization. 5 Whole exome sequencing has uncovered recurrent gains at 1q21, 6p22 and 11p13, loci that contain several genes (histones, IL6R, MCL1, and CD44) mechanistically linked with B-cell lymphomagenesis.7
MYC deregulation, due to rearrangement, amplification, or activation of certain signaling pathways (e.g., STAT3) is considered important for the development of PBL.11,17-19 The frequency of MYC abnormalities (rearrangement and/or gain) in our series (55%) was higher than that reported previously for PT-PBL (38%), but within the range (36-69%) reported for PBL arising in other settings.2,3,7,11,12,17,19 In contrast to the study of mostly HIV-related PBL by Valera et al., MYC rearrangements were more common in EBV– PT-PBL,19 and in some cases, the rearrangement was detected upon PTLD transformation to PBL or at disease relapse.
Recently, whole exome sequencing and targeted sequencing studies have unveiled the mutational landscape, primarily of HIV-related PBL,7,11 but until now the spectrum of genetic alterations underlying PT-PBL has not been explored. While HIV-related PBL and PT-PBL appear to exhibit overlapping genomic changes, some differences are evident.
Table 2.
Morphologic and immunophenotypic features of post-transplant plasmablastic lymphomas.
Figure 2.
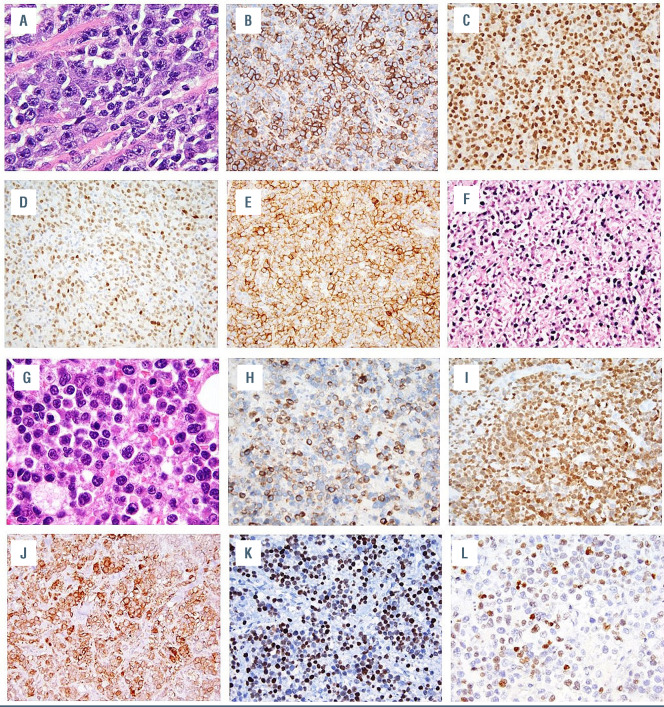
Immunophenotypic features of post-transplant plasmablastic lymphomas. (A) Hematoxylin and eosin (H&E)-stained section of tissue from case 2 showing plasmablasts, which are (B) partially positive for CD138 and (C) diffusely positive for MUM1 and display variable (D) PAX5 and (E) CD56 expression and evidence of (F) EBV infection by in situ hybridization for EBER. (G) H&E-stained section of case 9 showing plasmablasts, which are (H) partially positive for CD79a, (I) diffusely positive for MUM1 and display (J) aberrant CD10 expression, (K) P53 overexpression, and (L) moderate (30%) MYC expression.
In our series of PT-PBL, mutations in epigenetic modifiers were among the most frequent alterations (73%). Inactivating mutations of the KMT2/MLL family of histone H3 methyltransferases, which were most common, promote neoplasia via modification of global transcriptional activity.20 These mutations, particularly in KMT2D, have also been identified in monomorphic PTLD (DLBCL) (39%) and immunocompetent DLBCL (30-43%) and occur in 6-10% of cases of MM.21-26 Infrequent KMT2A mutations (6%), but no KMT2D mutations, have been reported in HIVrelated PBL.7 Recurrent loss-of-function mutations were also observed in the methylcytosine dioxygenase TET2 (27%), which have been shown to alter gene transcription via widespread DNA hypermethylation, a process important in the pathogenesis of PTLD.27 TET2 mutations are frequent in immunocompetent DLBCL and MM22,23,25,26,28 and occur in 9% of HIV-related PBL.7 In contrast to their common occurrence in EBV+ DLBCL, in our cohort TET2 mutations were exclusively seen in EBV– PBL.29 Other epigenetic modifiers recurrently mutated in HIV-related PBL include EP300, which was mutated in one PT-PBL, as well as TRRAP and HDAC6, which were not in our sequencing panel.7,11
Alterations (mutations and copy number changes) in DNA damage response and repair pathway genes were detected in 82% of our cohort, with chromosome 17/TP53 abnormalities being the most common recurrent events (55%). The frequency of TP53 mutations in PT-PBL (27%) was comparable to that reported in monomorphic PTLD (DLBCL) (36-44%) and, as in the latter, mutations, were more common in EBV– cases.21,30 A lower frequency of TP53 mutations has been observed in HIV-related PBL (9%).7 Of interest, TP53 mutations are present in up to 23% of immunocompetent DLBCL; however, a high proportion of DLBCL with plasmablastic/plasmacytoid features (85%) harbor TP53 deletions.24-26,31 TP53 mutations are uncommon in MM at diagnosis,22,23,28 but can be detected at disease progression, concomitant with TP53 deletions, and are associated with poor prognosis.23,32 MSI, resulting from mutations in DNA MMR genes, was identified in two EBV– PBL that showed a high mutation burden. MMR defects and MSI are unusual in B-cell non-Hodgkin lymphomas of immunocompetent individuals, but not infrequent in MM33 and immunodeficiency- associated B cell neoplasms, including PTLD.34
Table 3.
Genetic alterations in post-transplant plasmablastic lymphomas.
Gain-of-function mutations in MAPK pathway genes also appear to be more frequent in PT-PBL (55%) compared to HIV-related PBL (28%).7 Moreover, concurrent mutations of multiple MAPK pathway members, noted in several PT-PBL, could reflect the presence of multiple subclones, as described in MM.35 Different members of the MAPK pathway are mutated in diverse hematologic and lymphoid malignancies. However, KRAS and NRAS mutations, which are common in MM,23 are infrequent in immunocompetent DLBCL. Knowles et al. reported an NRAS mutation in a post-transplant plasmacytoid immunoblastic lymphoma, which could have represented a PBL.36 Well-known, activating BRAF K601E and codon 469 mutations were observed in our study. The latter and the canonical V600E mutation have also been documented in PBL arising in other settings.7,11 Intriguingly, the BRAF V600E mutation was recently reported in an immunomodulatory therapy-associated EBV+ anaplastic large cell lymphoma.37 Further studies are required to determine whether this mutation is a recurrent, lineageindependent, phenomenon in immune dysregulation-related lymphomas.
Recurrent mutations in members of the NOTCH signaling pathway, which controls B-cell fate determination,38 were observed in 45% of PT-PBL, mostly in EBV– cases (80% vs. 17% in EBV+ cases); it is unclear whether EBNA2 activates NOTCH signaling in a proportion of EBV+ PBL.39 Gain- and loss-of-function alterations in NOTCH pathway genes have been described in a variety of hemato-lymphoid neoplasms, including 24% of HIV-related PBL and some immunocompetent PBL.7,1 The pathogenesis of immunocompotent DLBCL of the N1 molecular subclass, which harbor NOTCH1 mutations and display a plasmacytic phenotype,26 is considered to be distinct from DLBCL of the BN2/Cluster 1 molecular subclass that have mutations in NOTCH2 and/or the NOTCH regulator, SPEN.24,26 However, we observed mutations in both, NOTCH1 and NOTCH2, as well as SPEN, at times concurrently, in PT-PBL. Deregulated activity of the NOTCH signaling pathway has also been implicated in the pathogenesis of MM, facilitating plasma cell growth and migration, but via different (non-mutational) mechanisms.40
JAK/STAT signaling, due to constitutive activation consequent to mutations, downstream effects of cytokine signaling, or EBV infection, contributes to the pathogenesis of several types of lymphoid neoplasms.41 Recurrent alterations in constituents of the JAK/STAT pathway, as observed in 36% of our cases and a higher proportion (62%) of HIV-related PBL, are known or predicted to enhance signaling. The STAT3 D661Y mutation, also detected in 8% of HIV-related PBL7 and STAT6 E372K mutations, occur in the SH2 and DNA binding domains, respectively, resulting in nuclear localization and activation of the transcription factors.42,43 Recurrent STAT3 mutations were noted exclusively in EBV+ HIV-related PBL,11 but mutations in several JAK/STAT pathway members, including STAT3, were observed in both EBV+ and EBV– PT-PBL. Multiple concomitant mutations in SOCS1, a negative regulator of JAK family proteins, present in one PT-PBL, have not been functionally characterized, but are predicted to inactivate SOCS1.44 Abrogation of SOCS1 and SOCS3 function by epigenetic silencing or mutations has been described in other immunodeficiency-associated non-Hodgkin lymphomas, including monomorphic PTLD (DLBCL) and polymorphic PTLD.45
Mutations in immune surveillance-associated genes also occurred in PT-PBL. FAS, a member of the tumor necrosis factor receptor superfamily and an important mediator of Tcell cytotoxicity, was recurrently mutated in our series. FAS mutations have not been previously reported in PTLD or PBL, but have been documented in MM.46 They are also common in immunocompetent DLBCL, particularly in Cluster 1, which frequently also harbor NOTCH pathway mutations,24 and, as in our series, are almost exclusively seen in EBV– cases.29 In addition to FAS mutation, one PBL had a frameshift mutation in CD58, another immune surveillancerelated protein required for activation of natural killer cells, which is commonly mutated in immunocompetent DLBCL.47
Prior studies have reported variants in PRDM1, an inducer of terminal B-cell differentiation and regulator of MYC, in 20-50% of PBL.11,17 However, many of the variants were not expected to be deleterious.11,17 PRDM1 variants were detected in 4% of HIV-related PBL.7 None of the PT-PBL in our cohort had pathogenic PRDM1 mutations and only a single VUS (Q586H) was observed.
Similar to our findings in PT-PBL, differences in the frequency or spectrum of genomic abnormalities between EBV+ and EBV– tumors have also been delineated in other types of B-cell PTLD.21,48-50 A lower mutation burden has been noted in EBV+ compared to EBV– monomorphic PTLD (DLBCL); this has been ascribed to the inherent oncogenic activity of the virus and, hence, a reduced requirement for proto-oncogene or tumor suppressor gene alterations.
EBV+ cases constituted 55% of our PT-PBL, a frequency not significantly different from previous studies of PT-PBL (67-79%) and intermediate between that reported for HIVrelated PBL (75-90%) and immunocompetent PBL (33-50%).2-4,7,12 Data regarding the EBV latency program in PBL have been conflicting. Ambrosio et al. reported a non-canonical EBV latency program i.e., partial expression of proteins characteristic of type II latency with simultaneous expression of lytic phase proteins in HIV+ and HIV– cases.10 Similarly, gene expression studies of HIV-related PBL have shown a much higher expression of the EBV lytic genes (BALF4 and BALF5) than canonical latency program genes, in most cases.7 Castillo et al., however, described latency I or III in most HIV-related EBV+ PBL, latency I in immunocompetent PBL, and predominantly latency III in PT-PBL.3 The vast majority of the EBV+ cases in our study displayed a latency II profile, similar to the observations of Morscio et al.,2 but different from those of Zimmerman et al., who reported mostly latency 0/I in PT-PBL.14
In our series, PBL exhibited morphologic and immunophenotypic heterogeneity, in line with prior observations.2,51 Almost half of the cases, including both EBV+ and EBV– cases, displayed minor foci of plasmacytic differentiation, concordant with the findings of other investigators.3,51 This feature was more common in PBL with chromosome 17/TP53 abnormalities. As reported for other B-cell PTLD,48 the majority (64%) of PT-PBL showed evidence of germinal center transit. Although the B-cell program is characteristically downregulated in PBL,2,3 a proportion of cases express B-cell antigens.3,52,53 Partial CD20 expression was observed in 27% of PT-PBL, a frequency similar to that reported for other types of PBL (23%).53 Over half of the PT-PBL showed variable PAX5 and/or CD79a positivity. Expression of CD79a has been documented in 45% of HIV-related and 68% of PT-PBL3 and PAX5 in 23-26% of mostly HIV-related PBL.52,53 In contrast to the findings of Montes-Moreno et al., who noted more frequent CD20 and/or PAX5 expression in EBV– PBL, a larger proportion of EBV+ PT-PBL (60%) was PAX5+.53 The two EBV– PAX5+ and CD79a+ PBL had highlevel MSI; otherwise, no differences in functional groups of mutations were apparent between cases expressing or lacking B-cell antigens. Since all patients with PTLD preceding PBL received rituximab, it is unclear whether the anti-CD20 antibody therapy was responsible for CD20 negativity of the PBL and/or promoted plasmablastic differentiation. Expression of CD56 and CD10, observed in a subset of PTPBL, has been reported more frequently in HIV-related and PT-PBL.2,3,53 Moreover, variability in the Ki-67 proliferation indices of our PT-PBL is in line with the findings of Morscio et al. (25-100% Ki-67 labeling in PT-PBL).2
PD-L1 expression, tumor infiltration by PD1+ T cells, and upregulation of genes related to immune escape, have been observed in EBV+ PBL7,8,11 and PTLD.54 PD-L1 expression by tumor cells was observed in subsets of EBV+ and EBV– PTPBL; the latter also harboring mutations in immune evasionrelated genes (FAS and CD58). PD-1 expression by tumor cells noted in one case has been previously reported in 5% of PBL.52
Thorough clinicopathological correlation is essential for resolving the differential diagnosis of PBL, which includes other neoplasms with plasmablastic features e.g., large B-cell lymphoma arising from HHV8-associated multicentric Castleman disease, primary effusion lymphoma, and ALK+ large B-cell lymphoma. Negative staining for HHV8 and ALK excluded these possibilities. Distinguishing between PBL and plasmablastic MM, however, can be difficult and a multimodal approach is required to make a correct diagnosis. Serum paraprotein analysis is not helpful as monoclonal proteins can be observed in some PBL patients,14 as was the case in our series. Importantly, none of the PT-PBL with bone marrow biopsies showed morphologic or immunophenotypic evidence of marrow involvement, all lacked bone (lytic) lesions on imaging as well as myelomarelated laboratory abnormalities, including hypercalcemia, and the vast majority occurred at mucosal sites, findings that do not support a diagnosis of MM.55 Furthermore, although some of the genetic abnormalities observed in PT-PBL overlapped with those of MM, they were detected in both EBV+ and EBV– PT-PBL and in HIV-related PBL.7,11 Furthermore, the overall complement of alterations differed from that of MM.
PT-PBL usually occur late after transplantation (median 96 months; range, 2-360 months)2,14 and are more frequent in males and in recipients of heart and kidney allografts,2,3,12,14 which was also true in our series. However, in contrast to a predominance of skin and lymph node involvement described previously,2,3 we noted a high frequency of intestinal disease (55%), with primary skin involvement observed in only 18% of patients. In addition to the oral cavity, the gastrointestinal tract is also a frequent site for HIVrelated PBL.3
Age, stage, and nodal involvement influence the prognosis of PBL, which is typically poor,12 although long survival, as observed for some of our PT-PBL patients, has been reported previously.2 In contrast to Zimmermann et al., we did not observe that patients with EBV– PT-PBL or PBL harboring MYC and/or IGH rearrangements had a worse prognosis.14 We could not determine any correlation between patients’ outcome and particular functional groups of mutations or mutational burden, although most had stage IV disease. Most patients in our cohort received lymphoma-directed therapies using conventional chemotherapy and/or radiotherapy. However two patients, including one who is still alive, also received bortezomib, a proteasome inhibitor frequently used to treat MM, which, in combination with lymphoma regimens, has been shown to be effective in treating PBL.3
In summary, our study is the first to investigate the genetic landscape of PT-PBL, revealing recurrent mutations in epigenetic modifiers and DNA damage response and repair, MAPK, JAK/STAT, NOTCH, and immune surveillance pathway genes. The observed genomic alterations overlap those reported for HIV-related PBL as well as subtypes of immunocompetent DLBCL and MM. Our findings reiterate the phenotypic heterogeneity of this rare type of PTLD, provide novel insights into PT-PBL biology and identify pathways amenable to targeted therapies.
Supplementary Material
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th edition, Volume 2. 2016. [Google Scholar]
- 2.Morscio J, Dierickx D, Nijs J, et al. Clinicopathologic comparison of plasmablastic lymphoma in HIV-positive, immunocompetent, and posttransplant patients: single-center series of 25 cases and meta-analysis of 277 reported cases. Am J Surg Pathol. 2014;38(7):875-886. [DOI] [PubMed] [Google Scholar]
- 3.Castillo JJ, Bibas M, Miranda RN. The biology and treatment of plasmablastic lymphoma. Blood. 2015;125(15):2323-2330. [DOI] [PubMed] [Google Scholar]
- 4.Li YJ, Li JW, Chen KL, et al. HIV-negative plasmablastic lymphoma: report of 8 cases and a comprehensive review of 394 published cases. Blood Res. 2020;55(1):49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CC, Zhou X, Taylor JJ, et al. Genomic profiling of plasmablastic lymphoma using array comparative genomic hybridization (aCGH): revealing significant overlapping genomic lesions with diffuse large B-cell lymphoma. J Hematol Oncol. 2009;2:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkozy C, Kaltenbach S, Faurie P, et al. Array-CGH predicts prognosis in plasma cell post-transplantation lymphoproliferative disorders. Genes Chromosomes Cancer. 2017;56(3):221-230. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Filip I, Gomez K, et al. Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK–STAT pathway. Blood Cancer Discov. 2020;1(1):112-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravelle P, Pericart S, Tosolini M, et al. EBV infection determines the immune hallmarks of plasmablastic lymphoma. Oncoimmunology. 2018;7(10):e1486950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman J, Gentles AJ, Sujoy V, et al. Gene expression analysis of plasmablastic lymphoma identifies downregulation of B-cell receptor signaling and additional unique transcriptional programs. Leukemia. 2015;29(11):2270-2273. [DOI] [PubMed] [Google Scholar]
- 10.Ambrosio MR, Mundo L, Gazaneo S, et al. MicroRNAs sequencing unveils distinct molecular subgroups of plasmablastic lymphoma. Oncotarget. 2017;8(64):107356-107373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Reyero J, Martinez Magunacelaya N, Gonzalez de Villambrosia S, et al. Genetic lesions in MYC and STAT3 drive oncogenic transcription factor overexpression in plasmablastic lymphoma. Haematologica. 2021;106(4):1120-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loghavi S, Alayed K, Aladily TN, et al. Stage, age, and EBV status impact outcomes of plasmablastic lymphoma patients: a clinicopathologic analysis of 61 patients. J Hematol Oncol. 2015;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dierickx D, Tousseyn T, Sagaert X, et al. Single-center analysis of biopsy-confirmed posttransplant lymphoproliferative disorder: incidence, clinicopathological characteristics and prognostic factors. Leuk Lymphoma. 2013;54(11):2433-2440. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann H, Oschlies I, Fink S, et al. Plasmablastic posttransplant lymphoma: cytogenetic aberrations and lack of Epstein- Barr virus association linked with poor outcome in the prospective German posttransplant lymphoproliferative disorder registry. Transplantation. 2012;93(5):543-550. [DOI] [PubMed] [Google Scholar]
- 15.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257-2317. [DOI] [PubMed] [Google Scholar]
- 16.Pang J, Gindin T, Mansukhani M, Fernandes H, Hsiao S. Microsatellite instability detection using a large next-generation sequencing cancer panel across diverse tumour types. J Clin Pathol. 2020;73(2):83-89. [DOI] [PubMed] [Google Scholar]
- 17.Montes-Moreno S, Martinez-Magunacelaya N, Zecchini-Barrese T, et al. Plasmablastic lymphoma phenotype is determined by genetic alterations in MYC and PRDM1. Mod Pathol. 2017;30(1):85-94. [DOI] [PubMed] [Google Scholar]
- 18.Sarosiek KA, Malumbres R, Nechushtan H, Gentles AJ, Avisar E, Lossos IS. Novel IL-21 signaling pathway up-regulates c-Myc and induces apoptosis of diffuse large B-cell lymphomas. Blood. 2010;115(3):570-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valera A, Balague O, Colomo L, et al. IG/MYC rearrangements are the main cytogenetic alteration in plasmablastic lymphomas. Am J Surg Pathol. 2010;34(11): 1686-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menter T, Juskevicius D, Alikian M, et al. Mutational landscape of B-cell post-transplant lymphoproliferative disorders. Br J Haematol. 2017;178(1):48-56. [DOI] [PubMed] [Google Scholar]
- 22.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339): 467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maura F, Bolli N, Angelopoulos N, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun. 2019;10(1):3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large Bcell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi D, Gaidano G, Gloghini A, et al. Frequent aberrant promoter hypermethylation of O6-methylguanine-DNA methyltransferase and death-associated protein kinase genes in immunodeficiency-related lymphomas. Br J Haematol. 2003;123(3): 475-478. [DOI] [PubMed] [Google Scholar]
- 28.Walker BA, Mavrommatis K, Wardell CP, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka K, Miyoshi H, Sakata S, et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virusassociated lymphomas. Leukemia. 2019;33(7):1687-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courville EL, Yohe S, Chou D, et al. EBVnegative monomorphic B-cell post-transplant lymphoproliferative disorders are pathologically distinct from EBV-positive cases and frequently contain TP53 mutations. Mod Pathol. 2016;29(10):1200-1211. [DOI] [PubMed] [Google Scholar]
- 31.Simonitsch-Klupp I, Hauser I, Ott G, et al. Diffuse large B-cell lymphomas with plasmablastic/ plasmacytoid features are associated with TP53 deletions and poor clinical outcome. Leukemia. 2004;18(1):146-155. [DOI] [PubMed] [Google Scholar]
- 32.Lode L, Eveillard M, Trichet V, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010;95(11):1973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velangi MR, Matheson EC, Morgan GJ, et al. DNA mismatch repair pathway defects in the pathogenesis and evolution of myeloma. Carcinogenesis. 2004;25(10):1795-1803. [DOI] [PubMed] [Google Scholar]
- 34.Capello D, Rossi D, Gaidano G. Post-transplant lymphoproliferative disorders: molecular basis of disease histogenesis and pathogenesis. Hematol Oncol. 2005;23(2):61-67. [DOI] [PubMed] [Google Scholar]
- 35.Lohr JG, Stojanov P, Carter SL, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowles DM, Cesarman E, Chadburn A, et al. Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders. Blood. 1995;85(2): 552-565. [PubMed] [Google Scholar]
- 37.Gru AA, Williams E, Junkins-Hopkins JM. An immune suppression-associated EBVpositive anaplastic large cell lymphoma with a BRAF V600E mutation. Am J Surg Pathol. 2019;43(1):140-146. [DOI] [PubMed] [Google Scholar]
- 38.Gu Y, Masiero M, Banham AH. Notch signaling: its roles and therapeutic potential in hematological malignancies. Oncotarget. 2016;7(20):29804-29823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kempkes B, Ling PD. EBNA2 and its coactivator EBNA-LP. Curr Top Microbiol Immunol. 2015;391:35-59. [DOI] [PubMed] [Google Scholar]
- 40.Colombo M, Galletti S, Garavelli S, et al. Notch signaling deregulation in multiple myeloma: a rational molecular target. Oncotarget. 2015;6(29):26826-26840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32(21):2601-2613. [DOI] [PubMed] [Google Scholar]
- 42.Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz M, Li H, Bernard D, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liau NPD, Laktyushin A, Lucet IS, et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. 2018;9(1):1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capello D, Gloghini A, Baldanzi G, et al. Alterations of negative regulators of cytokine signalling in immunodeficiency-related non- Hodgkin lymphoma. Hematol Oncol. 2013;31(1):22-28. [DOI] [PubMed] [Google Scholar]
- 46.Landowski TH, Qu N, Buyuksal I, Painter JS, Dalton WS. Mutations in the Fas antigen in patients with multiple myeloma. Blood. 1997;90(11):4266-4270. [PubMed] [Google Scholar]
- 47.Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of b2- microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20(6):728-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vakiani E, Basso K, Klein U, et al. Genetic and phenotypic analysis of B-cell post-transplant lymphoproliferative disorders provides insights into disease biology. Hematol Oncol. 2008;26(4):199-211. [DOI] [PubMed] [Google Scholar]
- 49.Rinaldi A, Capello D, Scandurra M, et al. Single nucleotide polymorphism-arrays provide new insights in the pathogenesis of post-transplant diffuse large B-cell lymphoma. Br J Haematol. 2010;149(4):569-577. [DOI] [PubMed] [Google Scholar]
- 50.Rinaldi A, Kwee I, Poretti G, et al. Comparative genome-wide profiling of posttransplant lymphoproliferative disorders and diffuse large B-cell lymphomas. Br J Haematol. 2006;134(1):27-36. [DOI] [PubMed] [Google Scholar]
- 51.Colomo L, Loong F, Rives S, et al. Diffuse large B-cell lymphomas with plasmablastic differentiation represent a heterogeneous group of disease entities. Am J Surg Pathol. 2004;28(6):736-747. [DOI] [PubMed] [Google Scholar]
- 52.Laurent C, Fabiani B, Do C, et al. Immunecheckpoint expression in Epstein-Barr virus positive and negative plasmablastic lymphoma: a clinical and pathological study in 82 patients. Haematologica. 2016;101(8): 976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montes-Moreno S, Gonzalez-Medina AR, Rodriguez-Pinilla SM, et al. Aggressive large B-cell lymphoma with plasma cell differentiation: immunohistochemical characterization of plasmablastic lymphoma and diffuse large B-cell lymphoma with partial plasmablastic phenotype. Haematologica. 2010;95(8):1342-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green MR, Rodig S, Juszczynski P, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ofori K, Soderquist C, Murty VV, et al. The clinical and pathological features of plasma cell myeloma post solid organ transplantation. Am J Hematol. 2020;95(12):1531-1541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.