Abstract
The Brucella AMOS PCR assay was previously developed to identify and differentiate specific Brucella species. In this study, an abbreviated Brucella AMOS PCR test was evaluated to determine its accuracy in differentiating Brucella abortus into three categories: field strains, vaccine strain 19 (S19), and vaccine strain RB51/parent strain 2308 (S2308). Two hundred thirty-one isolates were identified and tested by the conventional biochemical tests and Brucella AMOS PCR. This included 120 isolates identified as B. abortus S19, 9 identified as B. abortus strain RB51, 57 identified as B. abortus biovar 1, 15 identified as B. abortus bv. 2, 1 identified as B. abortus bv. 2 (M antigen dominant), 7 identified as B. abortus bv. 4, and 22 identified as B. abortus S2308 and isolated from experimentally infected cattle. The Brucella AMOS PCR correctly identified each isolate as RB51/S2308, S19, or a field strain of Brucella.
The Brucella AMOS PCR assay (2, 3) was developed at the National Animal Disease Center to identify and differentiate Brucella abortus, B. melitensis, B. ovis, and B. suis bacteria (AMOS is an acronym for the Brucella species identified). An abbreviated multiplex AMOS PCR assay was developed to differentiate B. abortus into three categories: field strains, vaccine strain 19 (S19), and vaccine strain RB51 and the RB51 parental strain, U.S. Department of Agriculture challenge strain 2308 (S2308) (3).
The abbreviated AMOS assay is based on the insertion of the genetic element IS711 at a unique chromosomal locus in B. abortus bv. 1, 2, and 4 and the double insertion of IS711 at a specific locus in B. abortus RB51 (2, 3). One PCR primer is anchored within the IS711 sequence, while the differentiating primers are localized in the unique chromosomal DNAs adjacent to the insertion. The primers were selected to amplify up to three products of different sizes. The primers amplify a 498-bp product present in B. abortus bv. 1, 2, and 4 plus two vaccine strains, and they also amplify a 364-bp product from B. abortus RB51. Identification of S19 is based on a PCR primer pair which amplifies a short sequence (178 bp) (3) of the eri gene (essential for erythritol catabolism), present in all Brucella strains except B. abortus S19 (4). Thus, the identification of S19 is based on the absence of amplification of this target.
The classical method of identifying the species and biovars of Brucella strains requires a minimum of 5 days. A PCR procedure can differentiate the vaccine strains from the field strains in 24 h and will provide useful, early information to regulatory officials. The purpose of this paper is to report the results of using the abbreviated Brucella AMOS PCR as a rapid screening test for B. abortus field strains and vaccine strains.
Isolates from tissue and milk samples submitted to the National Veterinary Services Laboratories and cultures submitted from state, federal, and university laboratories were identified to Brucella species and biovar level by the conventional methods (1). The following tests were performed on the isolates: growth in the presence of basic fuchsin (1:25,000 and 1:100,000), thionin (1:25,000 and 1:100,000), thionin blue (1:500,000), penicillin (5 U/ml), and erythritol (1 mg/ml and 2 mg/ml plus 5% bovine serum); urease and catalase activity; lysis by the Tbilisi phage; H2S production; and CO2 dependence. The dominant antigen was determined by the microagglutination test. Rough isolates were tested for susceptibility to rifampin in order to identify RB51 isolates (5).
The isolates were also tested by the abbreviated Brucella AMOS PCR. With a sterile inoculating loop, a small quantity of inoculum was suspended in 0.5 ml of 0.85% sterile saline. The cell suspension (2.5 μl) was added to 22.5 μl of the master mix consisting of 60 mM Tris-HCl (pH 9.0), 15 mM (NH4)2SO4, 1.5 mM MgCl2, 250 μM concentrations of each of the four deoxynucleoside triphosphates, 1 U of Taq polymerase, and five-primer cocktail (0.2 μM each), as previously described (3). The mixture was cycled 35 times through a regimen of 1.2 min at 95°C, 2.0 min at 55.5°C, and 2.0 min at 72°C under the conditions previously described (2). The amplified products were separated by electrophoresis in a 2.5% Metaphor agarose gel (FMC) in the presence of 0.5× Tris-borate-EDTA and were visualized by staining with ethidium bromide under UV light. The results were recorded by photographic methods.
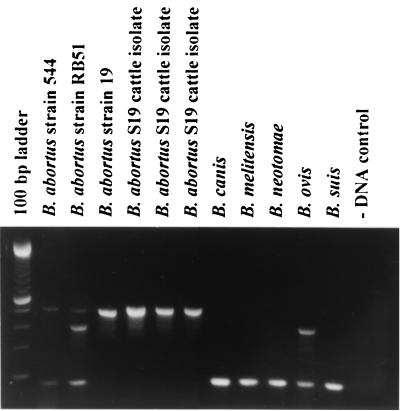
Figure 1 shows that the results for the B. abortus field strain, B. abortus vaccine strains, and representatives of the classical Brucella species were as expected. As designed, the typical B. abortus PCR amplifies two fragments, 498 and 178 bp. PCR using vaccine strain RB51 amplifies these two fragments plus the 364-bp fragment, while PCR with vaccine strain S19 produces only the B. abortus-specific 498-bp fragment. PCR with all other Brucella species except B. ovis amplifies only the 178-bp fragment from the eri gene. PCR with B. ovis, which has not been reported to infect cattle, also amplifies the 364-bp fragment associated with tandem copies of IS711, but this species is readily distinguished by the absence of the B. abortus-specific 498-bp fragment.
FIG. 1.
Amplification of DNA fragments from different Brucella strains. DNA was amplified by the abbreviated AMOS PCR assay described in the text. Eight microliters of amplicons was separated by electrophoresis, treated with ethidium bromide, and visualized under UV light. Strain 544 is biovar 1; B. abortus S19 in lane 4 is from a lab passage; B. abortus S19 in lanes 5 to 7 was reisolated from vaccinated cattle. For other species, the strains used were as follows: B. canis strain RM 6/66, B. melitensis bv. 1 strain 16M, B. neotomae strain 5K33, B. ovis strain 63/290, and B. suis bv. 1 strain 1330.
Two hundred thirty-one isolates from cattle and bison located in various states including Alabama, California, Florida, Kansas, Massachusetts, Texas, Vermont, Wisconsin, and Wyoming were identified and tested by the conventional biochemical tests and abbreviated Brucella AMOS PCR. This included 120 isolates identified as B. abortus S19, 9 identified as B. abortus strain RB51, 57 identified as B. abortus bv. 1, 15 identified as B. abortus bv. 2, 1 identified as B. abortus bv. 2 (M antigen dominant), 7 identified as B. abortus bv. 4, and 22 identified as B. abortus S2308. The 22 B. abortus S2308 samples had all been reisolated from experimentally challenged cattle. The abbreviated Brucella AMOS PCR was in 100% agreement with the conventional biochemical identification procedures in identifying the Brucella isolates tested (Table 1).
TABLE 1.
Results of the conventional identification method and the enhanced abbreviated Brucella AMOS PCR
| B. abortus biovar or strain | No. identified (classification) bya:
|
Lower 95% CI limit (%)b | |
|---|---|---|---|
| Conventional tests | PCR results | ||
| Biovar 1 | 57 | 57 (Field strain) | 94.9 |
| Biovar 2 | 16 | 16 (Field strain) | 82.9 |
| Biovar 4 | 7 | 7 (Field strain) | 65.2 |
| S19 | 120 | 120 (S19) | 97.5 |
| RB51 | 9 | 9 (RB51/S2308) | 71.7 |
| S2308 | 22 | 22 (RB51/S2308) | 87.3 |
| Total | 231 | 231 | 98.7 |
All results showed 100% agreement between the two methods.
CI, confidence interval.
The abbreviated Brucella AMOS PCR correctly identified each isolate as either RB51 or S2308, S19, or the field strain of Brucella. This PCR procedure has high potential as a rapid screening test for differentiating the two Brucella vaccines from the virulent field strains of Brucella. B. abortus strains RB51 and S2308 are not differentiated from each other by the abbreviated Brucella AMOS PCR. B. abortus strain S2308 is the U.S. Department of Agriculture challenge strain and is distinct from the virulent field strains in both its PCR pattern and biochemical characteristics. It also differs from RB51 because it forms smooth, colonies, while RB51 forms rough ones. B. abortus strain RB51 is resistant to rifampin while growth of the rest of the B. abortus strains is inhibited by rifampin.
In this study, PCR was shown to be a valuable tool for differentiating the vaccine strains from the field strains of Brucella. The conventional methods of identification require a minimum of 5 days to identify an isolate to Brucella species and biovar level. This can delay the movement of cattle between different owners and have a negative impact on the owners' financial planning. This study indicates that Brucellosis eradication program personnel could reliably use the abbreviated Brucella AMOS PCR to supplement other diagnostic and epidemiological data (such as herd history and serological test results) to release sale animals from quarantine before the conventional identification methods are completed.
REFERENCES
- 1.Alton G G, Jones L M, Angus R D, Verger J M. Techniques for the brucellosis laboratory. Paris, France: Institut National de la Recherche Agronomique; 1988. Bacteriological methods; pp. 13–61. [Google Scholar]
- 2.Bricker B J, Halling S M. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. 1994;32:2660–2666. doi: 10.1128/jcm.32.11.2660-2666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bricker B J, Halling S M. Enhancement of the Brucella AMOS PCR assay for differentiation of Brucella abortus vaccine strains S19 and RB51. J Clin Microbiol. 1995;33:1640–1642. doi: 10.1128/jcm.33.6.1640-1642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sangari F J, Garcia-Lobo J M, Agûero J. The Brucella abortus vaccine strain B19 carries a deletion in the erythritol catabolic genes. FEMS Microbiol Lett. 1994;121:337–342. doi: 10.1111/j.1574-6968.1994.tb07123.x. [DOI] [PubMed] [Google Scholar]
- 5.Schurig G G, Roop II R M, Bagchi T, Boyle S, Buhrman D, Sriranganathan N. Biological properties of RB51; a stable rough strain of Brucella abortus. Vet Microbiol. 1991;28:171–188. doi: 10.1016/0378-1135(91)90091-s. [DOI] [PubMed] [Google Scholar]