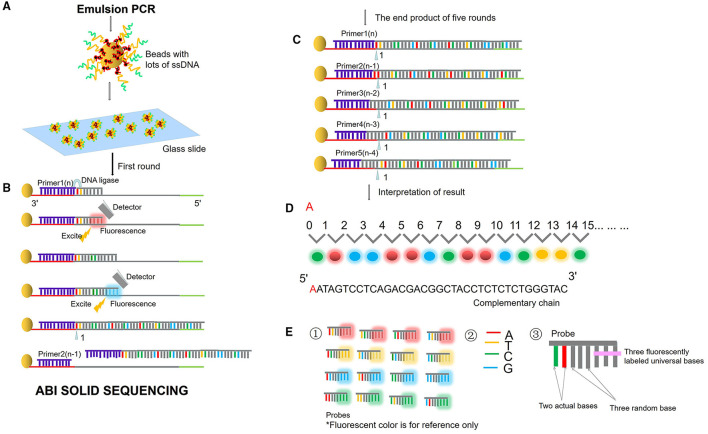
Figure 4.
ABI sequencing by oligonucleotide ligation and detection (SOLiD). (A) Emulsion PCR. After the emulsion is amplified, a large number of amplified products from the DNA template are attached to the magnetic beads. Then, the magnetic beads are deposited on the glass slide. (B) SLB. The first circle of sequencing reaction includes: the sequencing primer is combined with the template; the probe is combined with the template; the ligase connects the sequencing primer and the probe, excites the probe fluorescence, and removes the last three bases of the probe. Then plus the second probe, connects the two probes with the ligase, excites the probe fluorescence, and removes the last three bases of the probe. Repeat the above reaction to synthesize the whole chain. (C) Solid template shift. After cutting off the whole synthetic ligation, add sequencing primer n-1, repeat the above steps and start the second circle of reaction. Unidirectional solid sequencing requires five circles of sequencing reactions. The primers are n, n-1, n-2, n-3, and n-4, respectively, and the bases at each position are detected two times. (D) Fluorescent signal reception and interpretation of results. (E) Two-base encoding principle. ① “two-base encoding” specifies the corresponding relationship between different base pairs and four probe colors in the coding region (e.g., red fluorescence represents AT, TC, CA, GT). ② Four bases. ③ The probe is an 8-base single-chain fluorescent probe. The base pairs at positions first and second of the probe are determined, positions from third to fifth are random bases, which can be any of the four bases of “A, T, C, and G,” and positions from sixth to eighth mark four fluorescent dyes of different colors.