Summary
It has been a landmark year for artificial intelligence (AI) and biotechnology. Perhaps the most noteworthy of these advances was Google DeepMind’s AlphaFold2 algorithm which smashed records in protein structure prediction (Jumper et al., 2021, Nature, 596, 583) complemented by progress made by other research groups around the globe (Baek et al., 2021, Science, 373, 871; Zheng et al., 2021, Proteins). For the first time in history, AI achieved protein structure models rivalling the accuracy of experimentally determined structures. The power of accurate protein structure prediction at our fingertips has countless implications for drug discovery, de novo protein design and fundamental research in chemical biology. While acknowledging the significance of these breakthroughs, this perspective aims to cut through the hype and examine some key limitations using AlphaFold2 as a lens to consider the broader implications of AI for microbial biotechnology for the next 15 years and beyond.
For the first time in history, AI achieved protein structure prediction rivaling the accuracy of experimentally‐determined structures. Acknowledging the significance of this breakthrough, this perspective aims to cut through the hype and examine some key limitations using AlphaFold2 as a lens to consider the broader implications of AI for microbial biotechnology for the next 15 years and beyond.
A brief introduction
Given all the headlines surrounding AlphaFold2, it has been almost impossible for scientists to escape exposure to this major scientific breakthrough. Readers familiar with how AlphaFold2 works are encouraged to skip ahead. For those new to the topic, however, a brief introduction into AlphaFold2 architecture and why it is so groundbreaking is warranted. For a more in‐depth introduction, readers are referred to the original AlphaFold2 paper (Jumper et al., 2021), and other educational resources (EMBL‐EBI, 2021; Greener et al., 2021; Pereira and Alva, 2021).
In late 2020, results from the Critical Assessment of Protein Structure Prediction (CASP14) competition were announced. AlphaFold2, the brainchild of Google’s sister company, DeepMind, achieved a median backbone accuracy within the width of one carbon atom for protein structure prediction using primary amino acid sequences as inputs (Jumper et al., 2021). Scientists have been grappling with the protein folding problem for nearly half a century; yet, AlphaFold2 marked the first time that computational modelling achieved accuracy scores on par with experimental results. While there are many reasons behind the success of AlphaFold2, the meticulous engineering, including an iterative refinement mechanism and a new ‘attention’ architecture likely played a key role. Put simply, AlphaFold2’s architecture enables it to learn complex evolutionary sequence‐to‐structure relationships that simpler homology modelling methods cannot capture.
In a nutshell, AlphaFold2 starts with an input that most biologists are familiar with: a multiple‐sequence alignment (MSA) of the query sequence against evolutionarily related sequences. An important caveat is that for optimal performance, the MSA must consist of at least 30 sequences. Therefore, AlphaFold2 may underperform with inputs of unusual protein sequences with few homologues or de novo designed antibodies or proteins. The MSA is used to construct a ‘pair representation’ or contact map of which amino acids are likely to interact and build initial structural templates. The MSA and pair representation are then passed as inputs into a neural network with a Transformer architecture termed ‘Evoformer’. Transformer models were first pioneered in the field of natural language processing for sequence‐to‐sequence translation, such as Japanese‐to‐English or other language translation tasks. For AlphaFold2, the Transformer model is used to iteratively exchange information between the MSA and pair representations and refine them as inputs for another neural network, the ‘structure module’. The structure module predicts the 3D structure which is then iteratively fed back into Evoformer for several rounds of refinement until a final model is achieved. This is a vastly oversimplified summary and readers seeking more details are encouraged to consult the supplement of the original paper (Jumper et al., 2021). One of the major takeaways from the feat of AlphaFold2 is that there is no inherent magic involved. It is the combination of an impressive knowledge base, engineering prowess, and compute capabilities at Google’s DeepMind, now made available for the average user through a web interface (Mirdita et al., 2021).
Peering into the black box
The immensity of the accomplishment of AlphaFold2 is undeniable. The purpose of this perspective, however, is to go beyond the well‐deserved hype to examine the limitations and future implications. One of the greatest challenges facing AI is model interpretability. Neural networks are often referred to as black boxes due to complex transformations occurring in hidden layers. Although techniques, such as network deconvolution, exist to try to tease apart the underlying features, model interpretation remains a complicated process and active area of research. The power of AI presents a double‐edged sword in that non‐linear transformations in hidden layers enhance predictive power but simultaneously renders the transformed features uninterpretable to humans. Luckily, AlphaFold2 output (the coordinates for the 3D structural model of a protein) can easily be visualized in a way that is intuitive and interpretable for humans. Numerous resources are available to interpret results and the informative EMBL‐EBI training webinar is especially recommended (EMBL‐EBI, 2021). With AlphaFold2 now available for general users over web servers (Mirdita et al., 2021), it is more important than ever to understand what AlphaFold2 output means, where it excels and what the limitations are. A brief, non‐comprehensive summary of selected advantages and limitations are listed in Table 1.
Table 1.
Summary of selected advantages and limitations of AlphaFold2.
| Advantages | Limitations |
|---|---|
|
|
As with all AI applications, a healthy dose of scepticism is required when interpreting the output. For users viewing AlphaFold2 models, a useful piece of advice summed up succinctly by Dr. Jon Agirre at the University of York is, ‘Get rid of the spaghetti, trust the fusilli’. In other words, regions of AlphaFold2 models which are coloured orange or red indicate disordered ‘spaghetti’ regions which have a low level of prediction confidence relative to folded blue ‘fusilli’ regions. An example is shown in the AlphaFold2 model of titin (Fig. 1), the largest known human protein, visualized using the open‐source modelling software ChimeraX (Pettersen et al., 2021). This coloured distinction is a major advantage of AlphaFold2 since it indicates to users which regions of models can be trusted or not and helps guide experimental investigations.
Fig. 1.
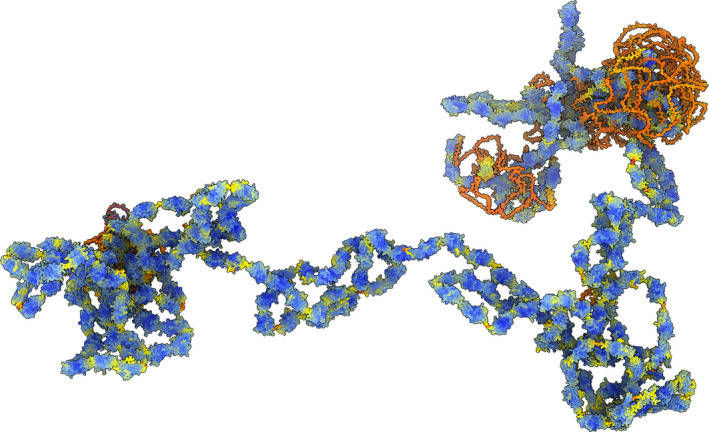
ChimeraX (Pettersen et al., 2021) visualization of human muscle protein, titin. Titin is 34,350 residues in length, and the model was produced by combining 29 segment structures, 1400 amino acids each, from the AlphaFold database. Details and code to reproduce the visualization available here: https://rbvi.github.io/chimerax‐recipes/big_alphafold/bigalpha.html, courtesy of Tom Goddard, ChimeraX.
Taking a broader perspective on the limitations, I argue that AI can assist with, but will generally fall short of, making truly novel biological discoveries without additional empirical validation. AI predictions are rooted in what is already known. To explore new regions of sequence space and characterize proteins of unknown function, experimentalists will continue to play a critical role. As an example, intrinsically disordered regions of proteins (IDPs) are estimated to encompass over 30% of sequences which are 30 amino acids or longer sampled from the human proteome (Necci et al., 2021). This estimate is now further bolstered by AlphaFold2 modelling of all human proteins (Tunyasuvunakool et al., 2021), with many regions of orange spaghetti overlapping with those regions that are predicted to be intrinsically disordered. In the case of IDPs and other regions of proteins that are highly flexible, classical techniques, such as nuclear magnetic resonance spectroscopy, will remain essential to track the range and timescales of protein dynamics. Cryo‐electron microscopy is another powerful method for structural analysis of macromolecular complexes which is a task where AlphaFold2 currently falls short. Overall, I anticipate one of the greatest barriers facing the advance of AI is its limited ability to predict the ‘unknown unknowns’.
AI and the reproducibility crisis
It is clear that AlphaFold2 and other AI advances have tremendous potential for biotechnology when users are aware of the limitations. But can AI also improve scientific reproducibility? Many of us bench scientists have experienced the gut‐wrenching moments when protocols that worked reliably for months suddenly and inexplicably produce a different result. The reality is that many experimental variables cannot be perfectly controlled. Aside from obvious human errors, the difference between a failed and successful experiment can come down to subtle discrepancies between laboratory personnel when pipetting or performing a certain technique. When moving from the bench to animal models or translational human studies, it is often even more challenging to reproduce results due to high costs and long timescales. This lack of reproducibility undermines credibility and leads to a distrust of science in the general public. Although the scientific process itself is rigorous and often inherently self‐correcting, AI can further help standardize the process through automation of experimental design, protocols, quality control and data analysis.
I optimistically predict that in the next 15 years and beyond, we will see a corresponding rise in experimental reproducibility as more steps in protocols use robotic rather than human arms and code workflows rather than copy‐and‐pasted spreadsheets. Added benefits include improved safety due to fewer accidents and fewer exhausted personnel carrying out long experiments by hand in the laboratory. Yet another bonus will be a reduction in common overuse injuries among laboratory personnel from performing repetitive tasks like pipetting. Nonetheless, a high level of biological variability will always plague experiments involving living organisms. In this context, the scalability of high‐throughput methods enabled by AI will provide higher statistical power to decipher biological signals through the noise. High‐throughput screening methods will, in turn, generate larger training datasets to feed into AI models and enable iterative design–build–test–learn cycles.
On the other hand, AI faces its own challenges with reproducibility. In a survey of artificial intelligence papers presented at major conferences, only 6% out of the 400 papers surveyed included complete, open‐source code (Hutson, 2018). In the case of AlphaFold2, the code was not made open source until nearly one year after the initial report, and the release was likely sped up by pressure from the scientific community. Open‐source code not only improves reproducibility, but also crowdsources the development process so that third‐party researchers can contribute to existing AI tools. The release of RoseTTAFold (Baek et al., 2021), with accuracy scores nearing AlphaFold, is one example of how community contributions are essential to keep the AI ecosystem accountable and healthy. Among a selection of other useful frameworks, the principles of Findability, Accessibility, Interoperability and Usability (FAIR) offer a solid foundation for data and code management across the disciplines of biology and AI (Wilkinson et al., 2016). In practice, this can be enforced through a combination of easy access to raw data and metadata, well‐documented code with good test coverage and version control and a web interface enabling access for programmers and non‐programmers alike. For more information on AI reproducibility, we refer readers to several articles (Stodden et al., 2016; Hutson, 2018; Haibe‐Kains et al., 2020).
In a widely cited survey of more than 1500 scientists on reproducibility, more than half identified ‘low statistical power’ or ‘specialized techniques that are difficult to repeat,’ as major causes of irreproducible results (Baker, 2016). As described above, automation enabled through AI can assist with both of these factors. What AI cannot assist with, however, is ‘pressure to publish’ and ‘selective reporting,’ which more than 60% of respondents said always or often contributed to irreproducibility. These latter two challenges are symptomatic of high‐stakes academic environments and poor mentor–mentee relationships, neither of which are likely to be solved by AI. This speaks to a broader truth that many come to recognize throughout their careers: it is not typically the technology, but rather the social problems that ultimately hamper progress.
Looking to the future: the next 15 years
As our ability to construct accurate structural models from primary sequence grows, a natural question is, what comes next? The potential applications of AlphaFold2 are only limited by one’s imagination. One area of active research is the leap from predicting protein structure to function. For applications in drug discovery and design, pre‐print servers are already overflowing with in silico studies using AlphaFold2 to model compound–protein, peptide–protein and protein–protein interactions (Bryant et al., 2021; Tsaban et al., 2021; Wang and Dokholyan, 2021). Basically any of the current limitations of AlphaFold2 (Table 1), including post‐translational modifications, multi‐domain complexes, intrinsically disordered regions and structural dynamics, are opportunities for new developments in the field. One particularly promising direction is improving the prediction of conformational dynamics, ranging from allosteric regulation of protein domains to the movement of individual active site residues. Ligand‐binding regions of proteins tend to be highly flexible and deviate from standard protein‐folding rules. These regions are also critical for determining protein function and substrate specificity and correspondingly, drug design. While AlphaFold2 performs remarkably well at overall structure prediction, the next level is to use AI to accurately predict the dynamics of key amino acid residues that move during interactions with a substrate.
Looking beyond AlphaFold2 to consider AI broadly, I envision a future in which AI enables enhanced reproducibility and predictability in biological experiments. Experimental systems involving living organisms rarely behave as expected. AI can remedy this by using design–build–test–learn cycles to train models to better predict and understand the natural world. This builds on the engineering principles established in the field of synthetic biology in which functional biological units are modelled, tested and characterized as components of larger living devices. AI also requires a shift in the scale of data from tens of data points to hundreds of millions of data points. High‐quality, large datasets are key to more reliably model and predict biological outcomes. Ultimately, this vision manifests itself as the coupling of ‘artificial’ AI engineering principles with the ‘natural’ sciences to open new frontiers for scientific exploration.
The human factor
For the future of AI for biotechnology, the continued importance of the human factor cannot be overemphasized. Many have hailed the rise of AI applications in biology as the beginning of a progression towards all biologists hunched over their computers rather than studying organisms in the laboratory or nature. Will biotechnology ever become a completely in silico discipline with experiments fully automated by robots? Some bench scientists have voiced fears that their jobs will be replaced by AI and their skill sets rendered useless. I would argue the opposite: that AI frees scientists from some of the more repetitive tasks to address larger questions in science. We experimentalists will always find curious new ways to get our ‘hands dirty’ at the bench or in the field. Experiments are especially important at the intrinsically disordered regions between scientific domains where we do not yet have contact maps of the key interactions. It is only through the combination of AI and the critical design of experiments by humans that scientific knowledge will advance. This sentiment is well summarized by Lindsey Backman, PhD candidate and HHMI Gilliam fellow at MIT, who writes about ‘the power of combining computational methods like Alphafold with traditional experimental methods like crystallography.’ Indeed, as Backman emphasizes, ‘it’s an exciting time to be a structural biologist.’
Funding information
S.L.R. is supported by Eawag discretionary funds.
Conflict of interest
The author has no conflict of interest to declare.
Acknowledgement
The author would like to acknowledge Jon Agirre and Lindsey Backman for their and contributions to the manuscript.
Microbial Biotechnology (2021) 15(1), 65–69
All quotes in this article were used with permission from the authors.
References
- Baek, M. , DiMaio, F. , Anishchenko, I. , Dauparas, J. , Ovchinnikov, S. , Lee, G.R. , et al. (2021) Accurate prediction of protein structures and interactions using a three‐track neural network. Science 373: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, M. (2016) 1,500 scientists lift the lid on reproducibility. Nature 533: 452–454. [DOI] [PubMed] [Google Scholar]
- Bryant, P. , Pozzati, G. , and Elofsson, A. (2021) Improved prediction of protein‐protein interactions using AlphaFold2 and extended multiple‐sequence alignments. bioRxiv. doi: 10.1101/2021.09.15.460468 [DOI]
- EMBL‐EBI. How to interpret AlphaFold structures (2021). Training webinar. URL https://www.ebi.ac.uk/training/events/how‐interpret‐alphafold‐structures/.
- Greener, J.G. , Kandathil, S.M. , Moffat, L. , and Jones, D.T. (2021) A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 10.1038/s41580-021-00407-0. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Haibe‐Kains, B. , Adam, G.A. , Hosny, A. , and Khodakarami, F. 2020) Transparency and reproducibility in artificial intelligence. Nature 586: E14–E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson, M. (2018) Artificial intelligence faces reproducibility crisis. Science 359: 725–726. [DOI] [PubMed] [Google Scholar]
- Jumper, J. , Evans, R. , Pritzel, A. , Green, T. , Figurnov, M. , Ronneberger, O. , et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita, M. , Ovchinnikov, S. , and Steinegger, M. (2021) ColabFold ‐ Making protein folding accessible to all. bioRxiv. doi: 10.1101/2021.08.15.456425 [DOI] [PMC free article] [PubMed]
- Necci, M. , Piovesan, D. , Predictors, C.A.I.D. , Curators, D.P. , and Tosatto, S.C.E. (2021) Critical assessment of protein intrinsic disorder prediction. Nat Methods 18: 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J. , and Alva, V. (2021) How do I get the most out of my protein sequence using bioinformatics tools? Acta Crystallogr D Struct Biol 77: 1116–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F. , Goddard, T.D. , Huang, C.C. , Meng, E.C. , Couch, G.S. , Croll, T.I. , et al. (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodden, V. , McNutt, M. , Bailey, D.H. , Deelman, E. , Gil, Y. , Hanson, B. , et al. (2016) Enhancing reproducibility for computational methods. Science 354: 1240–1241. [DOI] [PubMed] [Google Scholar]
- Tsaban, T. , Varga, J. , Avraham, O. , Ben‐Aharon, Z. , Khramushin, A. , and Schueler‐Furman, O. (2021) Harnessing protein folding neural networks for peptide‐protein docking. bioRxiv. doi: 10.1101/2021.08.01.454656 [DOI] [PMC free article] [PubMed]
- Tunyasuvunakool, K. , Adler, J. , Wu, Z. , Green, T. , Zielinski, M. , Žídek, A. , et al. (2021) Highly accurate protein structure prediction for the human proteome. Nature 596: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , and Dokholyan, N. V. (2021) Yuel: Compound‐protein interaction prediction with high generalizability. bioRxiv. 10.1101/2021.07.06.451043 [DOI] [PMC free article] [PubMed]
- Wilkinson, M.D. , Dumontier, M. , Aalbersberg, I.J.J. , Appleton, G. , Axton, M. , Baak, A. , et al. (2016) The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]


