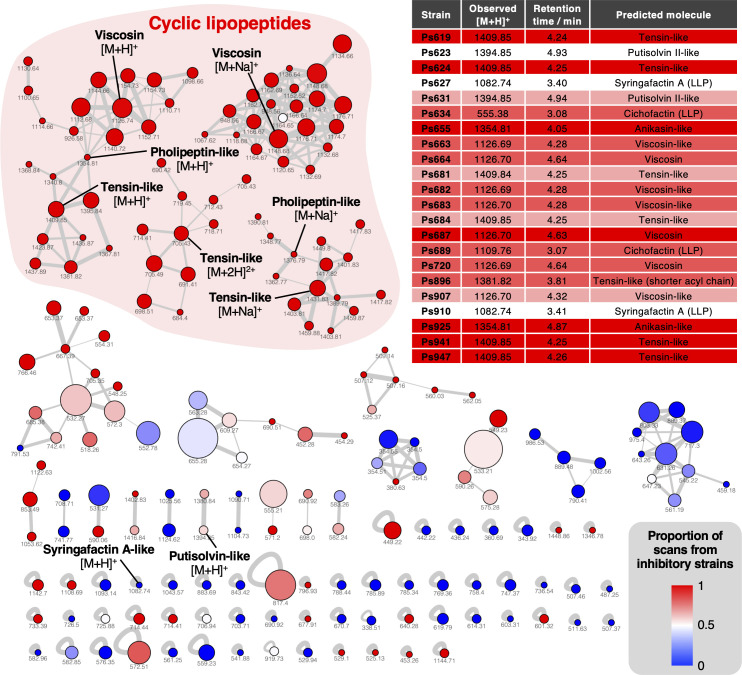
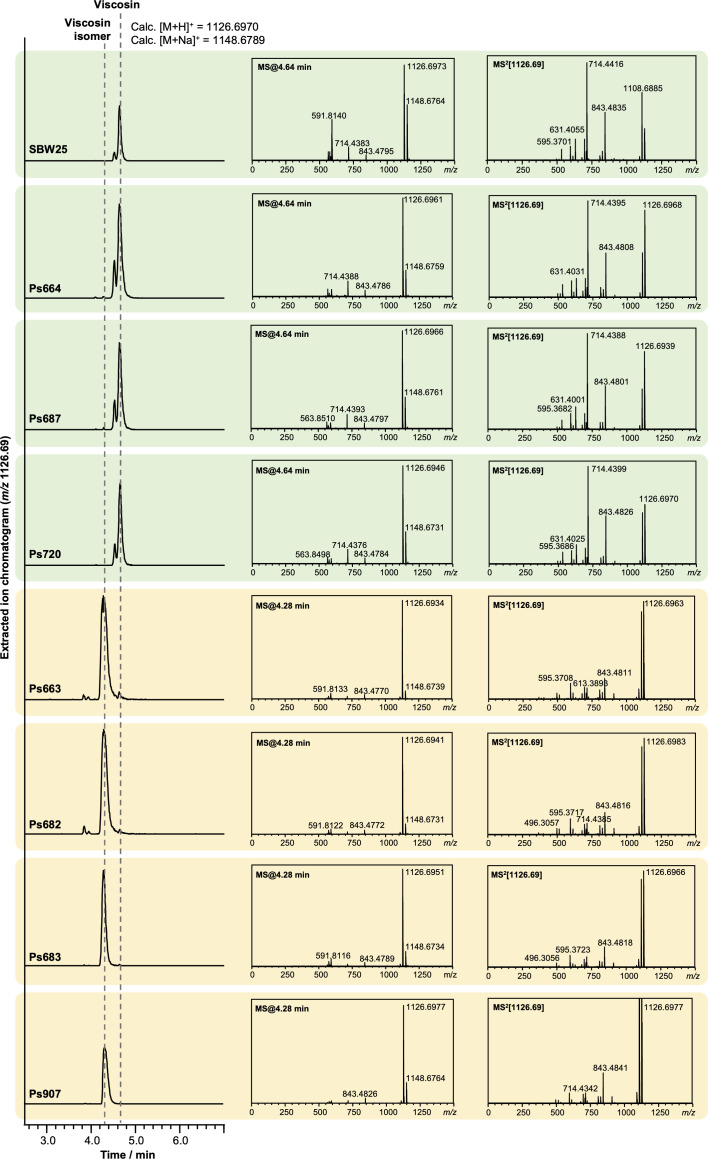
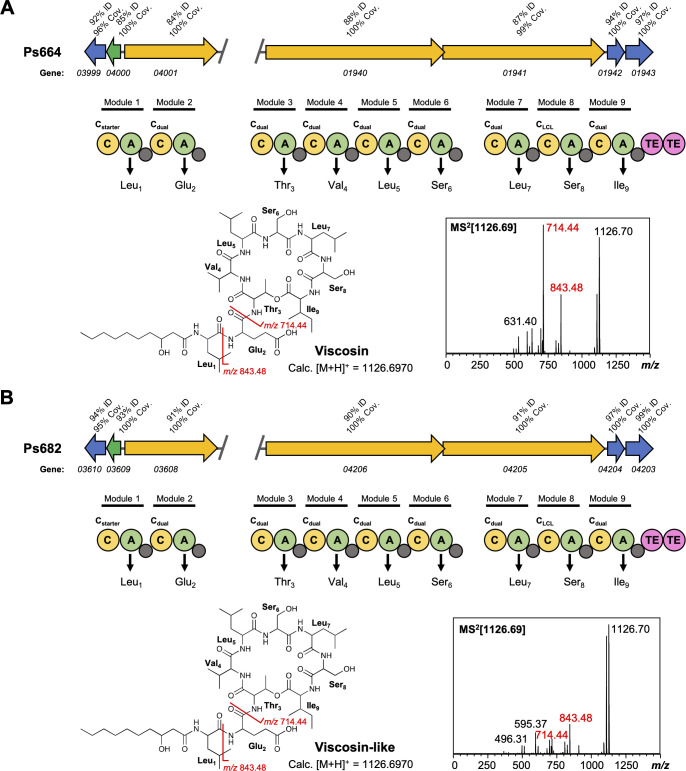
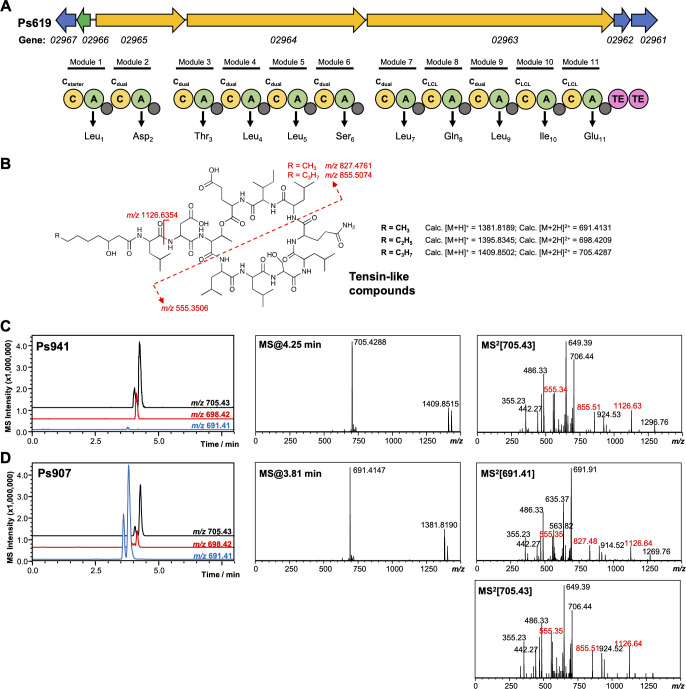
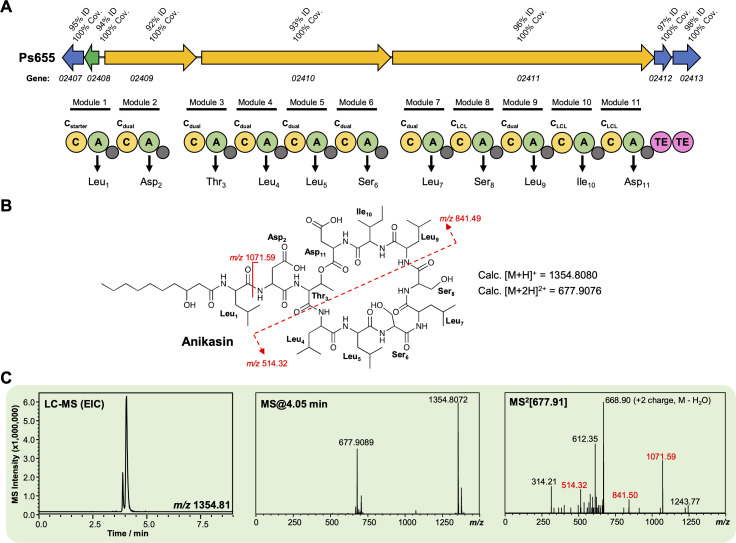
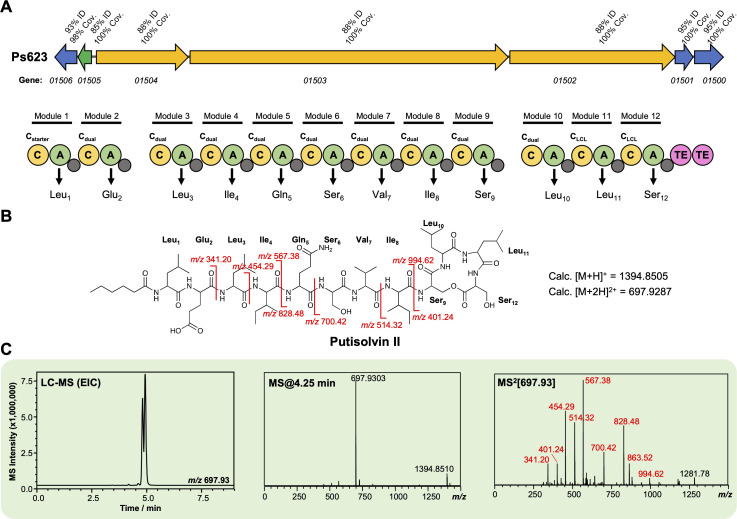
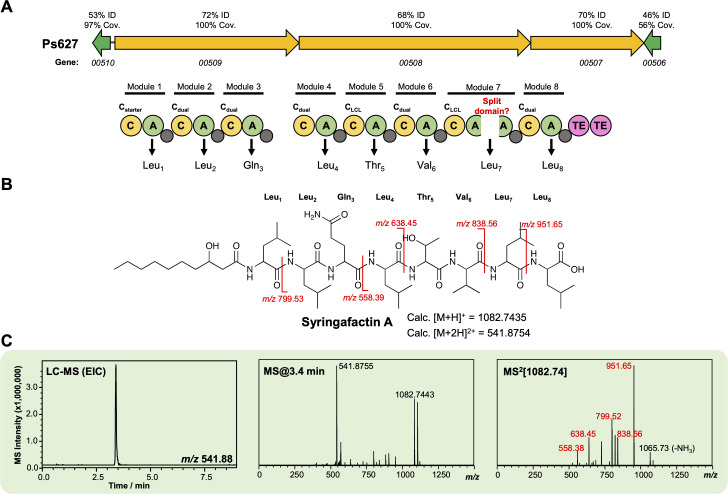
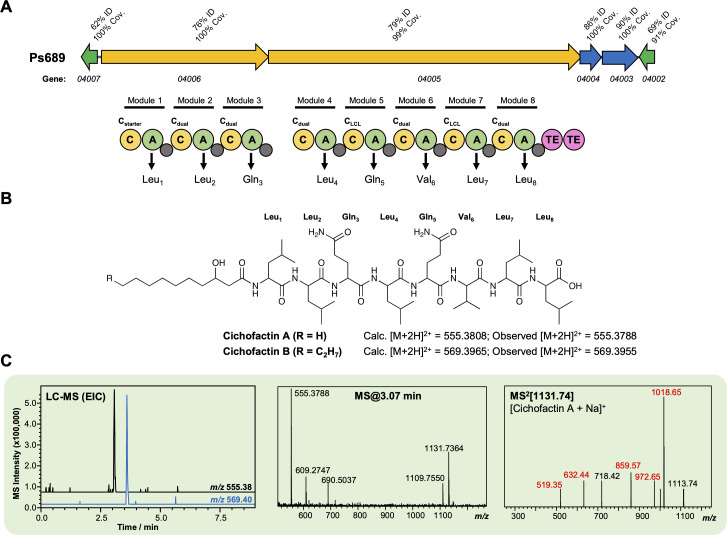
Figure 4. Mass spectral networking analysis of liquid chromatography–tandem mass spectrometry (LC-MS/MS) data from the Pseudomonas strains used in this study.
Node area is proportional to the number of distinct strains where MS/MS data were acquired for a given metabolite. Node color reflects the proportion of MS/MS scans for a given node that come from strains with a S. scabies inhibition score ≥1. Nodes are labeled with the corresponding parent masses and nodes that relate to lipopeptides are labeled (multiple networks arise from differential fragmentation of [M + H]+, [M + 2H]2+, and [M + Na]+ ions). Line thickness is proportional to the cosine similarity score calculated by Global Natural Product Social Molecular Networking (GNPS) (Aron et al., 2020). The table shows production of lipopeptides by strains containing lipopeptide biosynthetic gene clusters (BGCs). Color coding reflects level of S. scabies inhibition by each strain with same scale as Figure 2 (LLP: linear lipopeptide; all others are cyclic lipopeptides [CLPs]).