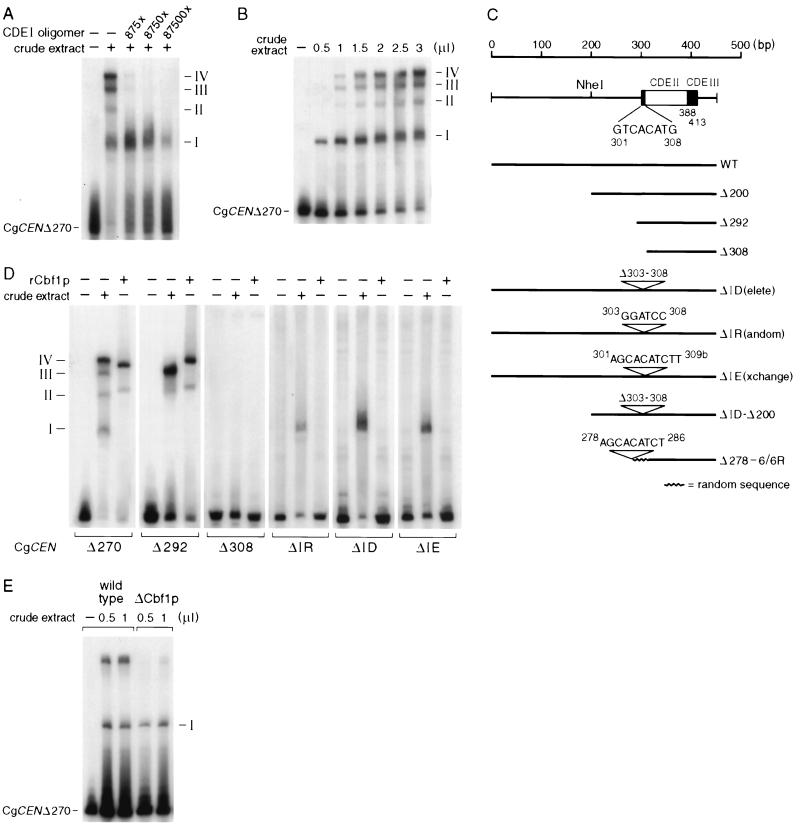
FIG. 4.
Gel mobility shift assays with C. glabrata crude extracts and a labeled CgCEN DNA fragment. Shown are autoradiograms of protein-DNA complexes that were formed after incubation of CgCEN DNA with C. glabrata crude extracts and the results of fractionation on nondenaturing gels as described previously (35). (A) Specific competition of complex formation with a CDEI oligomer. Radioactively labeled CEN DNA (5 pmol) was incubated with 2 μl of C. glabrata crude extract in the presence or absence of increasing amounts of an unlabeled, concatemerized CDEI binding site. The numbers on the right indicate the four different complexes that were observed with wild-type CEN DNA. (B) Concentration dependence of protein-DNA complex formation. The same amount of radiolabeled CEN DNA used to obtain the results in panel A was incubated with increasing amounts of crude extract. (C) Centromere mutants that were tested in bandshift experiments and in vivo plasmid loss assays (Fig. 5). WT, wild type. (D) Protein-DNA complex formation on mutated centromere DNAs. Centromere DNA fragments were generated by PCR and radiolabeled as described in Materials and Methods. Centromere DNAs (5 pmol) were incubated with either 2 μl of crude extract or with a 1:20 dilution of purified rCbf1p. (E) Protein-DNA complex formation of CgCbf1p-depleted cells. A crude extract made from C. glabrata 98CBF1 cells (Fig. 8) grown for 7 h in the presence of 10 g of doxycycline per ml was incubated with CEN DNA.