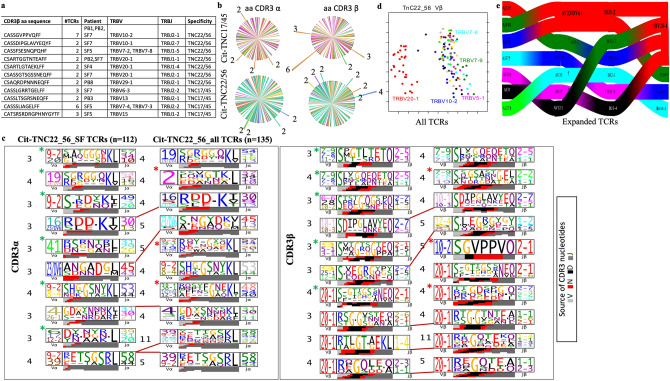
Figure 3.
Features of cit-TNC specific TCR repertoire in RA: (a) Sharing of amino acid sequences in the TCR-CDR3 beta was found between 3 different patients and within 7/8 patients. Shared TRBV and TRBJ gene usage patterns are shown in the context of the different patient samples and citrullinated antigens. Only cit-TNC22/56 specific CDR3 sequences showed sharing between patients. (b) Sharing of exact CDR3α (left column) or CDR3β (right column) amino acid sequences between different TCRs as shown for TNC17/45 sequences (upper row), and TNC22/56 (bottom row). The protruding arms in the figure represent shared TCRs where each color signifies a different sample. (c) The figure shows CDR3 consensus sequences (logos) obtained using TCRdist, where amino acids are marked according to their source genes. The first panel shows the comparison of patterns in the CDR3 alpha chain of cit-TNC 22/56 specific repertoire in SF (left column) and all (SF + PB) samples (right column). The second panel shows the patterns in CDR3 beta sequences. For both alpha and beta, the patterns shared between PB and SF of different samples are marked with connecting lines, while SF restricted and PB restricted patterns are marked in green and red asterisks respectively. (d) Principal component analysis (PCA) of TRBV gene usage between all cit-TNC22/56-specific TCRs shows differential distribution of TRBV20-1 gene (red) while other gene segments display considerable overlap. (e) Paired alpha/beta TCR gene usage patterns of cit-TNC22/56 specific expanded TCRs from the four patients are shown on the right. The gene usage pattern (alpha on the left and beta to the right) show genes in order of their prevalence (top color representing maximum proportion). TRBV20-1 (left side in red) was the most used BV gene and paired gene-usage patterns showed similarities among different cit-TNC22/56-specific TCRs from different RA patients.