Figure 2: Exome-wide analysis of germline variant discovery in the presence and absence of additional genomics datasets.
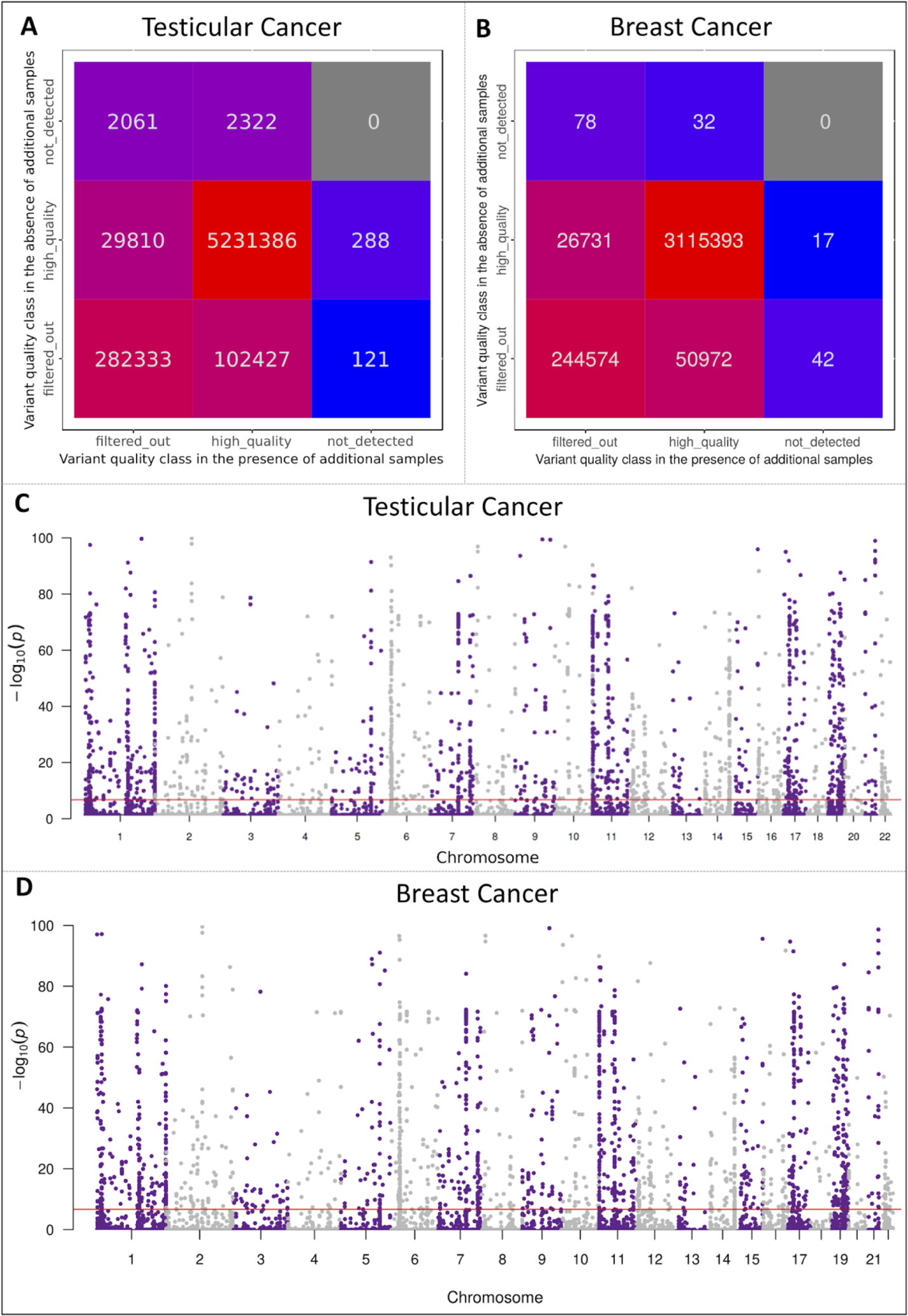
A and B; Confusion matrices of the final quality classification status of the germline variants detected in the testicular and breast cancer cohorts, respectively, between the first and second computational runs. C and D; Manhattan plots of the p-values for the germline variants, filtered by GATK-JG in both computational runs, to be absent by chance in a randomly selected 239 individuals from the European ancestry. A total of 184,827 variants had a p-value <1.76e-07 (depicted in 2C by the horizontal dotted red line) in the testicular cancer cohort and 116,078 variants had a p-value <2.04e-07 (depicted in 2D by the horizontal dotted red line) in the breast cancer cohort, suggesting a non-random underdetection effect of the GATK-JG for common variants across coding regions.
