Abstract
Problem:
Mucosal-Associated Invariant T (MAIT) cells have been recently identified at the maternal-fetal interface. However, transcriptional programming of decidual MAIT cells in pregnancy remains poorly understood.
Method of Study:
We employed a multiomic approach to address this question. Mononuclear cells from the decidua basalis and parietalis, and control PBMCs, were analyzed via flow cytometry to investigate MAIT cells in the decidua and assess their transcription factor expression. In a separate study, both decidual and matched peripheral MAIT cells were analyzed using Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) coupled with gene expression analysis. Lastly, decidual MAIT cells were stimulated with E.coli and expression of MR1 by antigen presenting cells was measured to evaluate decidual MAIT cell function.
Results:
First, we identified MAIT cells in both the decidua basalis and parietalis. CITE-seq, coupled with scRNA-seq gene expression analysis, highlighted transcriptional programming differences between decidual and matched peripheral MAIT cells at a single cell resolution. Transcription factor expression analysis further highlighted transcriptional differences between decidual MAIT cells and non-matched peripheral MAIT cells. Functionally, MAIT cells are skewed towards IFNγ and TNFα production upon stimulation, with E.coli leading to IFNγ production. Lastly, we demonstrate that MR1, the antigen presenting molecule restricting MAIT cells, is expressed by decidual APCs.
Conclusion:
MAIT cells are present in the decidua basalis and obtain a unique gene expression profile. The presence of MR1 on APCs coupled with in vitro activation by E.coli suggests that MAIT cells might be involved in tissue-repair mechanisms at the maternal-fetal interface.
Keywords: MAIT cells, Decidua Basalis, Decidua Parietalis, MR1, scRNA-seq, CITE-seq
1. Introduction
The maternal-fetal interface, specifically the decidua basalis and the decidua parietalis, is a critically important site for the growth and development of the fetus. Proper interplay between immune cells and non-immune cells (endothelial and trophoblasts) is required for tolerance of the semi-allogenic fetus and simultaneous protection against infection (1,2). Recently Mucosal-Associated Invariant T (MAIT) cells have been identified at the maternal-fetal interface (3–5), adding a layer of complexity to the decidual immunome. Despite these advances, however, transcriptional imprinting of decidual MAIT cells and their function, remain questions of pivotal importance to their impact on normal and pathological pregnancies.
MAIT cells are an innate-like T cell subset that expresses a T cell receptor (TCR) composed of an invariant Vα7.2+ chain (TRAV1–2 combined with either TRAJ33/12/20) coupled to a β-chain of restricted repertoire diversity (6). Unlike conventional T cells, MAIT cells are restricted by the monomorphic MHC Class I-like molecule MR1 (7–10). First identified in the intestinal lamina propria (7), MAIT cells have since been described in various tissues including the liver (11,12), lung (13,14), and cervix and endometrium (15).
The effector properties of MAIT cells have been well documented to be against microbial pathogens (16–18), specifically in response to microbial insults, MAIT cells produce IFNγ, TNFα, IL-17A, and IL-22 as well as cytotoxic molecules, Granzyme B and Perforin (18) in a tissue-specific manner (15,17,19,20). Tied to their function, MAIT cells primarily express transcription factors PLZF, RORγt, and T-bet (6,21) while Eomes expression has been noted in MAIT cells in certain conditions (15,22).
Despite their known presence at the maternal-fetal interface, questions remain as to whether decidual MAIT cells owe their phenotypic characteristics to their tissue residence. Here, we have leveraged recent advances in single cell technologies and taken a single cell multiomics approach to highlight specific features of decidual MAIT cells. We show that there is a specific transcriptional imprint in decidual MAIT cells compared to peripheral MAIT cells, both from pregnant and non-pregnant individuals. Furthermore, we identify multiple MR1-expressing antigen presenting cells (APCs) in the decidual, suggestive of MAIT cells activation through TCR engagement. Lastly, we show that decidual MAIT cells preferentially produce IFNγ and TNFα upon activation. Overall, our data supports the presence of MAIT cells in the decidua (3,4) and that decidual MAIT cells are unique from their PBMC counterparts.
2. Materials and Methods
2.1. Sample Collections and Processing
Placental samples were sourced from term normal elective cesarean sections (>37 wks GA) in accordance with the UW Obstetrical Tissue Bank IRB protocol (#2014–1223) and UnityPoint Health – Meriter IRB protocol (#2017–004). Decidua basalis tissue was dissected from the maternal surface of the placenta and decidua parietalis tissue was scraped from the fetal membrane as previously described (23). Decidual tissue was mechanically and enzymatically dissociated as previously described (24,25). Isolated mononuclear cells (MCs) were frozen in FBS/10% DMSO and stored until needed. Peripheral blood mononuclear cells (PBMC) technical controls (contPBMCs), from batched, anonymous, non-pregnant, reproductive age females, were purchased (All Cells®, Alameda, CA) and kept frozen until processing.
In a separate set of experiments, five individual placental and matched whole blood specimens were collected for scRNA-seq (CITEseq) under the approval of Meriter IRB# 2018–10 protocol. Specimens were obtained from five healthy women undergoing normal elective cesarean sections (>37 wks GA) on day of intake. Decidual samples were processed as described above. Matched PBMCs (matPBMCs) were isolated from whole blood via density gradient centrifugation. PBMC layer was extracted and washed with PBS, counted, adjusted to an optimal concentration of 4 X106 cells/mL and cryopreserved as described above.
2.2. T-Cell Activation
T cells were activated with either PMA/Ionomycin (Leukocyte Activation Cocktail with GolgiPlug™, BD Bioscience) or E.coli strain K12. For PMA/Ionomycin activation, MCs were treated for 6 hours with Leukocyte Activation Cocktail and monensin. After which, MCs were harvested and prepared for cell labeling.
E.coli stimulation was performed as previously described (26). Briefly, frozen E.coli aliquots (4X109 CFU/mL) were thawed and fixed in 1% Paraformaldehyde for 4 mins at room temperature. E. coli cell pellets were then washed three times with PBS before being resuspended in complete RPMI (109 CFU/mL). MCs were then incubated with fixed E.coli at a 10:1 CFU/PBMC multiplicity of infection (MOI). MCs were then stimulated by E.coli for 18 hours, with the addition of monensin and Brefeldin A for the last 6 hours of stimulation.
2.3. Cell Labeling
MCs were labeled with either Zombie NIR™ Fixable Viability Kit (BioLegend) or LIVE/DEAD® fixable blue stain (Invitrogen, Waltham, MA, USA) following manufacturer’s instructions. MCs were subsequently labeled with fluorochrome-conjugated monoclonal antibodies, listed in Table 1. Transcription factor assessment was performed using BD Pharmingen™ Transcription Factor Buffer Set, with overnight fixation. Cytokine production was assessed by first labeling MCs with cell surface antibodies followed by fixation (BD Cytofix™ Fixation Buffer). MCs were then permeabilized (BD Perm/Wash™ buffer) and intracellular targets were labeled, according to manufacturer instructions.
Table 1.
Fluorescent conjugated antibodies used for flow cytometry
| Marker | Clone | Fluorochrome | Supplier |
|---|---|---|---|
| CD3 | SK7 | PE-Cy7 | BD Science |
| CD3 | UCHT1 | BV421 | BD Science |
| CD3 | UCHT1 | PE | BioLegend |
| CD4 | SK3 | BUV496 | BD Science |
| CD4 | RPA-T4 | A488 | BD Science |
| CD8 | RPA-T8 | BV421 | BD Science |
| CD8 | SK1 | BV605 | BD Science |
| CD11b | ICRF44 | BV605 | BD Science |
| CD11c | B-ly6 | BB515 | BD Science |
| CD14 | MOP9 | BV510 | BD Science |
| CD14 | M5E2 | PE-Cy7 | BioLegend |
| CD16 | 3G8 | BUV496 | BD Science |
| CD19 | SJ25C1 | BV510 | BD Science |
| CD19 | SJ25C1 | PE-Cy7 | BD Science |
| CD27 | M-T271 | PE-Cy7 | BD Science |
| CD34 | 581 | PE-Cy5 | BD Science |
| CD45 | 2D1 | A488 | BioLegend |
| CD45RO | UCHL1 | APC-H7 | BD Science |
| CD45RO | UCHL1 | PE | BioLegend |
| CD56 | B159 | PE-Cy7 | BD Science |
| CD56 | NCAM16.2 | BV421 | BD Science |
| CD80 | L307.4 | A700 | BD Science |
| CD123 | 7G3 | BUV395 | BD Science |
| CD141 | 1A4 | APC | BD Science |
| CD161 | DX12 | PE-Cy5 | BD Science |
| CD163 | GHI/61 | PE-Dazzle594 | BioLegend |
| CD209 | DCN46 | PerCP-cy5.5 | BD Science |
| Eomes | WD1928 | PE-eFluor610 | eBiosciences |
| GATA-3 | L50–823 | BUV395 | BD Science |
| Granzyme B | GB11 | A647 | BioLegend |
| HLADR | G46–6 | BV786 | BD Science |
| IFN𝛾 | 4S.B3 | A488 | BioLegend |
| IL-17A | BL168 | PE-Dazzle594 | BioLegend |
| IL-22 | 22URTI | PerCP-eFluor 710 | eBiosciences |
| IL-22 | 2612A41 | APC | BioLegend |
| MR1 | 26.5 | PE | BioLegend |
| PLZF | Mags.21F7 | A488 | eBiosciences |
| ROR𝛾t | AFKJS-9 | APC | eBiosciences |
| Tbet | O4–46 | BV650 | BD Science |
| TCR V⍺7.2 | 3C10 | BV785 | BioLegend |
| TCR V⍺7.2 | 3C10 | BV510 | BioLegend |
| TNF⍺ | MAb11 | BV650 | BioLegend |
| TNF⍺ | MAb11 | BV785 | BioLegend |
Surface and intracellular expression levels of MR1 was assessed by labeling MCs with anti-MR1 during the surface labeling step (surface only) or after fixation/permeabilization step (Total: surface + intracellular). To account for any effects of fixation on fluorochrome brightness, both sets of samples were processed using the BD Pharmingen™ Transcription Factor Buffer Set as described above.
2.4. Flow Cytometry and Data Analysis
Samples were acquired using the ThermoFisher Attune NxT or the BD LSR Fortessa with 4 laser (405, 488, 561, 642nm), 14-detector configuration and 5 laser (355, 405, 488, 562, 633nm) 20-detector configuration, respectively. SPHERO™ Rainbow Calibration Particles (Spherotech, Lake Forest, IL, USA) were used to standardize PMT voltage settings as previously described (24).
Data was analyzed using FlowJo™ v.10 software (FlowJo LLC, Ashland, OR, USA) and statistical software GraphPad Prism (ver. 8, GraphPad Software Inc, La Jolla, CA, USA). Statistical significance was determined by Student t-test or One-way ANOVA where appropriate.
2.5. Single Cell RNA sequencing and data analysis
2.51. T cell Enrichment and Sorting
T cells from decidua basalis and matPBMCs were enriched using the Dynabeads® Untouched™ Human T Cells Kit (Invitrogen). T cell enriched samples were then labeled with Zombie NIR™ Fixable Viability Kit (BioLegend). Subsequently, cells treated with TruStain FcX™ (BioLegend), then labeled with fluorochrome- and TotalSeq™-C oligonucleotide-conjugated antibodies (Table 1 and 2). Samples were then sorted into PBS (Ca++/MG++ free, 1% non-acetylated BSA, 1 μL/mL RNase inhibitor) using the BD FACSAria in a 3 laser (488nm, 561nm, 405nm) configuration, collecting 100,000 live, CD3+ cells. A post-sort purity check was performed demonstrating a >98.5% purity (Figure S3).
Table 2.
Oligo conjugated antibodies used for CITE-seq
| Marker | Clone | Supplier |
|---|---|---|
| TotalSeq™-C TCR V𝛼7.2 | 3C10 | BioLegend |
| TotalSeq™-C CD161 | HP-3G10 | BioLegend |
| TotalSeq™-C CD4 | RPA-T4 | BioLegend |
| TotalSeq™-C CD8 | SK1 | BioLegend |
| TotalSeq™-C TCRV𝛼24-J𝛼18 | 6B11 | BioLegend |
| TotalSeq™-C TCR𝛼𝛽 | IP26 | BioLegend |
| TotalSeq™-C CD279 | EH12.2H7 | BioLegend |
| TotalSeq™-C CD196 | G034E3 | BioLegend |
| TotalSeq™-C CD194 | L291H4 | BioLegend |
| TotalSeq™-C CD197 | G043H7 | BioLegend |
| TotalSeq™-C CD45RA | HI100 | BioLegend |
2.52. Library preparations and sequencing
Cell suspensions were submitted to UW Biotechnology Center where cell viability was further validated using the Countess™ II (Invitrogen). A total of 56,000 cells (7000 per sample) were targeted using the 10X Genomics. Samples meeting all quality control criteria were selected for single cell RNA library preparation and sequencing. Libraries were prepared and downstream processing utilizing Illumina sequencing technology was performed by the University of Wisconsin Gene Expression Center in collaboration with the UWBC DNA Sequencing Facility, Madison, Wisconsin.
2.53. Clustering, Differential Expression, and CITE-seq data analysis
We used the CellRanger v3.1 count pipeline to generate filtered gene count matrices for each sample. This pipeline includes demultiplexing, discriminating between cell and background barcodes, and aligning reads to the human transcriptome (GRCh38 3.0). We next enriched the dataset for high quality cells by filtering on several quality control metrics: the fraction of UMIs aligning to mitochondrial transcripts, fraction of housekeeping genes detected (list from (27)), number of RNA or surface features detected, and number of RNA (FB) UMIs. We converted each value to median absolute deviations (MADs) within each sample and removed cells for which any value was outside of [−3,3]. Finally, although the cells were selected for CDR3 positivity prior to sequencing, we implemented two additional quality control filters to ensure a high confidence data set: (1) we required cells to have full-length CDR3 sequence data (from scVDJ-seq; unpublished results) and (2) we manually gated the cells using the CD4 and CD8 antibody features to identify and remove possible double-positives and double-negatives (of which very few remained after the CDR3 sequence requirement filter). At the end of the filtering process, both samples from one of the subjects had lost substantially more cells than the other three subjects. After removing both samples for that subject, we had three subjects each with PBMC and decidua basalis samples.
Count normalization.
For RNA values, we log-transformed the depth-normalized counts. For surface proteins profiled by feature barcoding, we used centered log-ratio (CLR) normalization across all cells for each feature.
Batch correction for visualization and clustering.
We used the Seurat v3 integration pipeline (28) to align the RNA datasets to reduce subject- and localization-specific variability. Seurat employs “anchor” cells, which are cells that have similar nearest neighborhoods in the datasets being reconciled. The corrected expression value for a gene in a cell is a function of its proximity to the anchor cells and a score for each anchor. First, we integrated the PBMC T cells across subjects, and separately the decidua basalis T cells across subjects. Then we integrated the PBMCs and decidua basalis cells together. The final batch-corrected RNA matrix was used for UMAP plots and clustering, but not for expression visualization or statistical analysis.
Cluster analysis to define the MAIT cell subset
To define T cell subsets, we used Seurat v3.1.5 to cluster the cells in the integrated RNA space (cluster resolution parameter=0.5). We examined the CITE-seq protein and RNA expression profiles of each cluster. One cluster contained 307 cells and uniquely exhibited high expression of proteins TCR-Vα7.2 and CD161. We called this the MAIT cluster.
Differential expression analysis
We sought to identify decidua basalis- and PBMC-specific genes within the MAIT cell cluster. Genes were filtered in advance of testing for expression in at least 10% of the cluster’s cells and for absolute average log fold change >0.25. To account for differences in the number of cells available from each subject, we first tested each gene within each subject separately using rank-sum tests on the log-normalized RNA counts. Next, for each gene, we combined the p-values from the three subjects using the sum-log method. To control for multiple testing, we applied Bonferroni correction to each combined p-value. For the denominator we used the number of genes with any nonzero expression in any cell in the dataset (33548 genes). Finally, differentially expressed (DE) genes were selected based on a threshold of adjusted p<0.05.
CITE-seq visualization.
Visualizations of single cell CITE-seq data were generated using the R packages Seurat (v3.1.5), ggplot2 (v3.3.2), and viridis (v0.5.1).
3. Results
3.1. Mucosal Associated Invariant T (MAIT) are present in Human Term Decidua
We first asked whether MAIT cells are present at the maternal-fetal interface. To address this question, we developed a 13-parameter panel (Table 1) to determine the abundance of MAIT cells in human term decidua and their phenotypes. We employed anonymous contPBMCs for quality control purposes and panel validation (Figure S1).
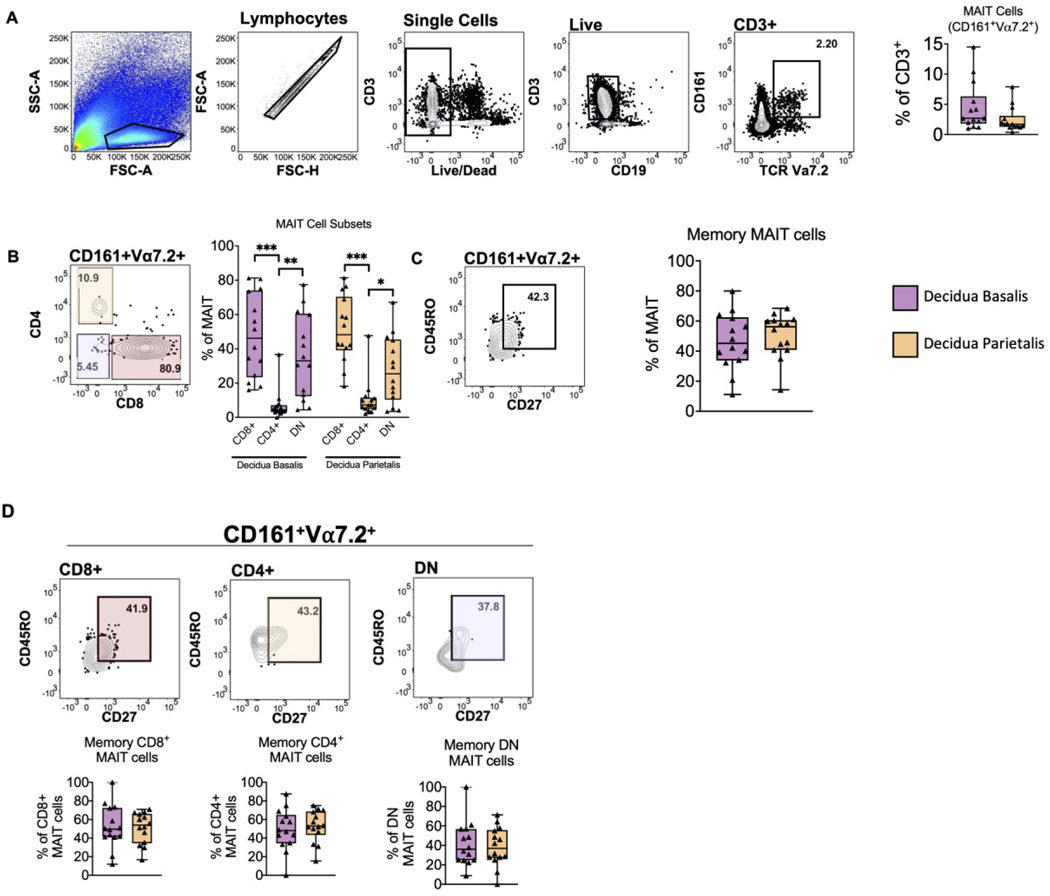
First, we found that the proportion of MAIT cells from total T cells was similar between the decidua basalis and parietalis (Figure 1A). Further analysis revealed a higher proportion of CD8+ MAIT cells, followed by DN cells, with a lower proportion of CD4+ MAIT cells (Figure 1B), a trend that we observed in both the decidua basalis and parietalis. Finally, we assessed decidual MAIT cells for a memory phenotype (CD161+Vα7.2+CD45RO+CD27+) and found that memory MAIT cells were present at a frequency between 40–60% of total MAIT cells in decidua basalis and parietalis (Figure 1C), with similar distributions of CD8+, CD4+ and DN memory MAIT cells in both the decidua basalis and parietalis (Figure 1D).
Figure 1. Mucosal Associated Invariant T (MAIT) cells identified in Human Term Decidua.
(A) Gating strategy identifying MAIT cells and frequency of MAIT cells in term decidua basalis and parietalis. (B) Identification of MAIT cell CD4+/CD8+ subsets (C) Identification of memory MAIT cells (CD45RO+CD27+) and (D) memory CD8+, CD4+, DN MAIT cell subsets. Decidua Basalis, n = 14; Decidua Parietalis, n = 14. Data represented as max/min, median, and 25 and 75th percentiles. Statistical significance was determined by Student’s t-test or One-way ANOVA, followed by Tukey post-hoc tests, were appropriate. *p < 0.05, **p < 0.005, ***p < 0.001.
Next, we validated the MAIT cell phenotype utilizing a tetramer-specific approach. Using the 5-(2-oxopropylideneamino)-6-D-ribitylaminouraci-l (5-OP-RU) loaded MR1 tetramer (Table 2), we confirmed that decidual MAIT cells were bound by the MR1:5-OP tetramer compared to the non-specific MR1:6-FP tetramer (Figure S2A). Overall, our results confirm the presence of MAIT cells in both term decidua basalis and parietalis, with most being CD8+/DN. Furthermore, we found no significant differences in MAIT cell composition between the decidua basalis and parietalis.
3.2. Single cell CITE-seq analysis reveals unique gene expression signature of decidual MAIT cells
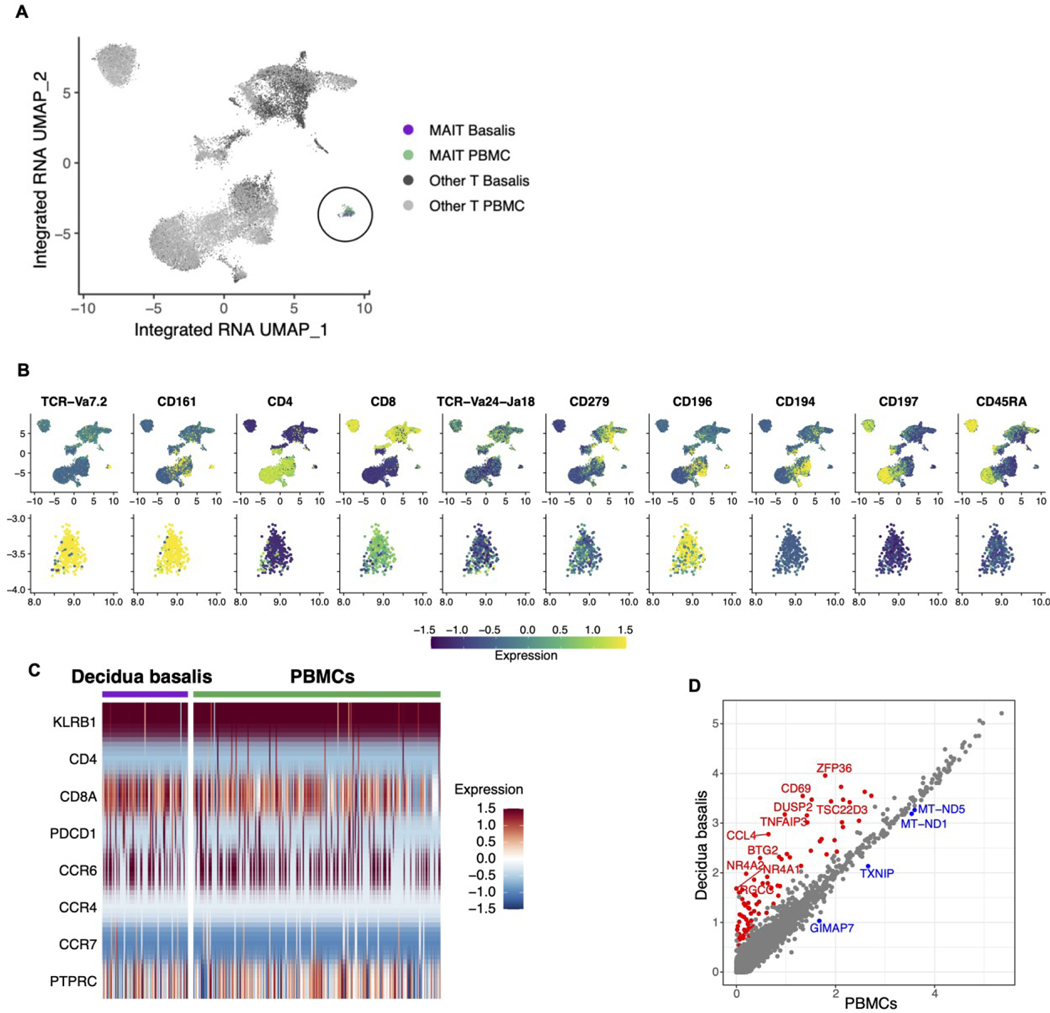
To better understand the influence of the decidual environment on MAIT cells, we employed single-cell RNA-seq analysis on both decidual and circulating MAIT cells. Using the Chromium Single Cell sequencing platform (10X Genomics), coupled with CITE-seq (29), allowing for simultaneous assessment of select protein expression, we obtained high-quality sequence profiles for 22,245 T cells T cells from both the decidua and matched PBMCs (on average 2,700 per decidua and 4,700 from matched PBMCs).Clustering analysis revealed a unique MAIT cell cluster, separate from other T cells (Figure 2A), consisting of 307 cells. MAIT cell phenotype was confirmed by CITE-seq analysis, showing the expression of MAIT-specific T cell receptor chain, Vα7.2, but not NKT-specific Vα24-Jα18 (Figure 2B). We further confirmed that cells within the MAIT cell cluster expressed CD161 (Figure 2B), a defining marker of MAIT cell identity (11,30,31). As expected, most MAIT cells identified expressed CD8, as opposed to CD4, lacked expression of CD279 (PD-1), CD194 (CCR4), CD197 (CCR7) and CD45RA, and express CD196 (CCR6) all suggesting an effector-memory phenotype (Figure 2B). Protein level expression was mirrored by gene expression levels (Figure 2C), with the notable exception of CD45RA, an isoform of CD45 (PTPRC).
Figure 2. Transcriptome analysis reveals unique gene expression profile of decidual MAIT cells.
(A) UMAP clustering of total T cells showing the segregation of the MAIT cell cluster with a total of 307 cells. (B) Expression of surface proteins profiled by CITE-seq overlaid on UMAP plots. CLR-normalized counts were scaled across all T-cells. UMAP plots were computed from a batch-corrected RNA matrix. Top row: all T cells. Bottom row: zoom in to MAIT cluster only. (C) Expression of genes corresponding to surface markers profiled by CITE-seq (B) in MAIT cells from decidua basalis and PBMCs. Log-normalized RNA expression was scaled across all T cells. (D) Average log-normalized RNA expression in MAIT cells found in PBMCs and decidua basalis. Each point is one gene. Color denotes statistically significant differential expression determined by combining p-values from per-subject Wilcoxon rank sum tests and applying a threshold of Bonferroni-adjusted p<0.05. Red: gene is significantly up-regulated in decidua basalis MAITs versus PBMC MAITs; blue: gene is significantly down-regulated; gray: not significant. The top ten up-regulated genes and all four down-regulated genes are labeled, ranking by average fold change.
Next, we asked whether the gene expression profile of decidual MAIT cells was different from matPBMC MAIT cells and found that there were differentially expressed genes between the two groups (Figure 2D). Notably, we found genes involved in immune suppression Zfp36 (32), Tnfaip3 (33), Dusp2 (34), Tsc22d3 (35), Btg2 (36), Nr4a2 (37), and Nr4a1(38), cell cycle regulation (Rgcc) (39), resident memory (Cd69) (40,41), and cell migration(Ccl4) (42) to be upregulated in decidual MAIT cells (Figure 2D). Conversely, genes involved in apoptosis (Gimap7) (43), glucose metabolism (Txnip) (44), and oxidative phosphorylation (Mt-nd1, Mt-nd5) (45) were downregulated in decidual MAIT cells (Figure 2D). Overall, these results confirm that presence of MAIT cells in term decidua and that decidual MAIT cells maintain a unique gene expression profile compared to their peripheral counterparts.
3.3. Decidual MAIT cell transcription factor expression analysis reveals surprising expression levels of Eomes and T-bet
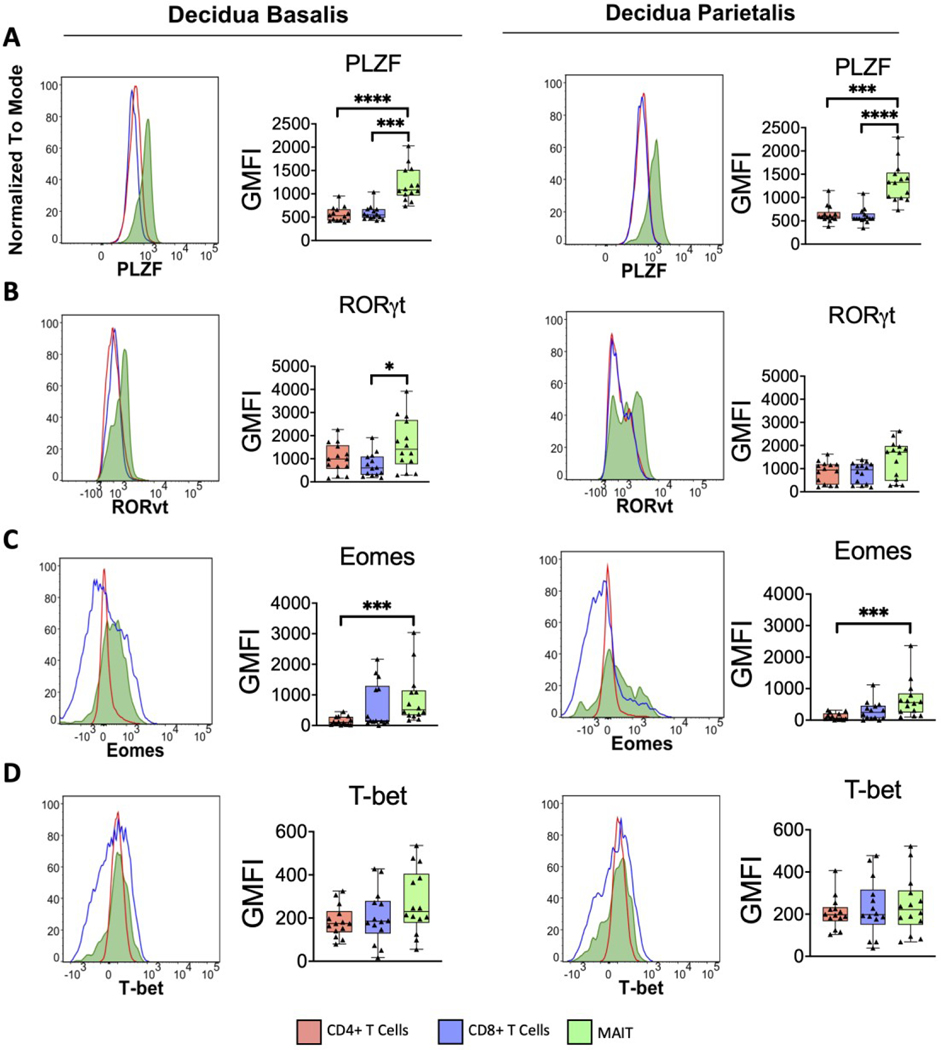
To better understand the transcriptional programming of decidual MAIT cells, we analyzed their expression of lineage-defining transcription factors (Figure 3). Promyelocytic leukemia zinc finger (PLZF) is recognized as a necessary transcription factor for the development of innate T cells (46). As such, we expected higher PLZF expression in decidual MAIT cells compared to their conventional T cell counterparts (Figure 3A).
Figure 3. Transcriptional profile of decidual MAIT cells sets them apart from decidual T cells.
Histograms showing range of expression of transcription factors (A) PLZF, (B) RORγt, (C) Eomes, and (D) T-bet for both decidua basalis and parietalis MAIT and conventional T cells. Plots of the geometric mean fluorescence intensity (GMFI) for each transcription factor are shown adjacent to corresponding histograms. Decidua Basalis, n = 14; Decidua Parietalis, n = 14. Data represented as max/min, median, and 25 and 75th percentiles. Statistical significance was determined by One-way ANOVA, followed by Tukey post-hoc tests. *p < 0.05, ***p < 0.001, ****p< 0.0001.
RORγt, the canonical lineage-defining transcription factor of TH17 and ILC3s, is tied to the production of IL-17 and IL-22 (47). Because previous reports have shown that MAIT cells are capable of producing TH17-type cytokines (15,20), we measured the level of RORγt expression in decidual MAIT cells. Higher levels of RORγt expression was observed in MAIT cells from the decidua basalis, compared to CD4+ and CD8+ T cells from the same tissue. However, we found no difference in the decidua parietalis across T cell subsets (Figure 3B).
Eomesodermin (Eomes) and T-bet, both members of the T-box transcription factor family, are involved in the development and function of various immune cells (48) and are known drivers of IFNγ expression (48–50). Because MAIT cells have the capacity of producing IFNγ (3,4,13,16,51,52), we asked whether they expressed Eomes and T-bet (Figure 3C&D). We found that decidual MAIT cells expressed higher levels of Eomes compared to decidual conventional T cells (Figure 3C). However, the same pattern was not observed in T-bet expression (Figure 3D). Taken together, decidual MAIT cells express different levels of lineage-defining transcription factors compared to conventional T cells, including higher levels of PLZF, RORγt, and Eomes.
Because we did not find any difference in transcription factor expression between decidua basalis and parietalis MAIT cells (Figure S4B), despite having different tissue origins, we asked if this observation was pregnancy specific. First, we compared decidual MAIT cells to contPBMCs (collected from non-pregnant females). We found that all transcription factors (PLZF, RORγt, Eomes, Tbet) were expressed at higher levels in contPBMC MAIT cells compared to decidual MAIT cells (Figure S4B). Because our matched samples were reserved for scRNA-seq analysis, we looked at transcript abundance to compare the expression level of these same targets in matched MAIT cells and found no difference between decidual and circulating MAIT cells (Figure S5). This suggests that location, either decidua or circulation, does not alter lineage-defining transcription factor expression within the same individual.
3.4. MR1 Antigen Presenting Cells (APCs) point to possible MAIT cell function in human term decidua
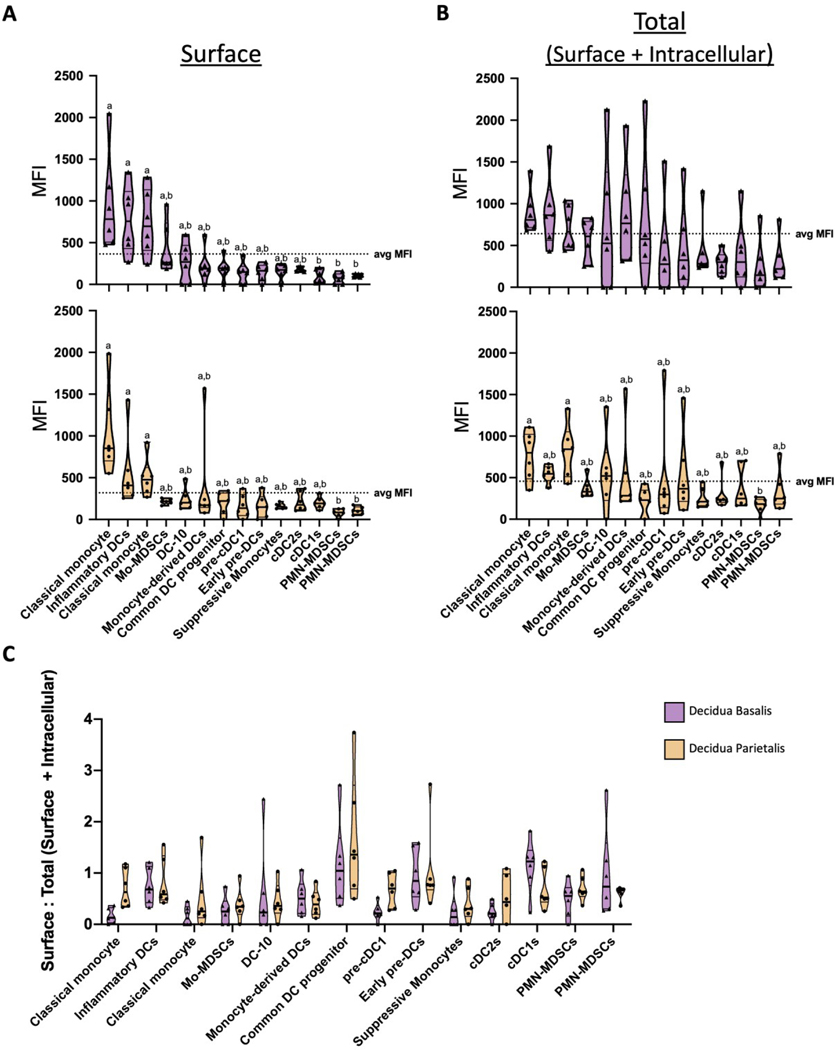
Canonical activation of MAIT cells occurs through the engagement of antigen via MR1 presentation (8,10). To determine if activation of decidual MAIT cells could occur in an MR-dependent fashion, we assessed the expression of MR1 on multiple antigen presenting cells (APCs) of the maternal-fetal interface (Figure S5; Table S1).
Because MR1 is expressed at low levels at the surface in the absence of a ligand (53), we measured both surface and intracellular expression (Figure S5). We first assessed surface expression of MR1 by APCs, in both the decidua basalis and parietalis (Figure 4A). The median fluorescence intensity (MFI) of MR1 was measured across multiple decidual APCs and found variable levels of MR1 expression (Figure 4A). Specifically, we found that classical monocyte 1, Inflammatory DCs, and classical monocyte 2 expressed higher levels of surface MR1 compared to classical type 1 DCs, PMN-MDSCs 1, and PMN-MDSCs 2 in decidual APCs (Figure 4A). Interestingly, when assessing total MR1 expression (surface and intracellular), only classical monocyte 1 and classical monocyte 2 from the decidua parietalis expressed higher levels of MR1 compared to PMN-MDSCs 1 (Figure 4B).
Figure 4. MR1 is expressed by antigen presenting cells (APCs) in term human decidua.
(A) Decidual antigen presenting cells (APCs) from both the decidual basalis (top) and parietalis (bottom) were labeled with anti-MR1 antibody and median fluorescence intensity (MFI) was calculated to assess expression. (B) To account for intracellularly expressed MR1, APCs were labeled both for surface and intracellular MR1 protein. (C) Surface to Total (Surface + Intracellular) MFI ratio across APCs in decidua basalis and parietalis was calculated. Decidua basalis, n = 6; Decidua parietalis, n = 6. Data represented as violin plots displaying max/min, median, and 25 and 75th percentiles. Statistical significance was determined by One-way ANOVA, followed by Tukey post-hoc tests. Different letters indicate statistical differences between groups (p < 0.05).
To better understand the proportion of surface versus intracellular levels of MR1 expression in decidual APCs, we calculated the ratio of surface to total (surface + intracellular) MR1 expression for both the decidua basalis and parietalis (Figure 4C). Most APCs had a ratio of less than 1, with the exception of Common DC progenitor and classical type 1 DCs, suggesting a greater amount of intracellular MR1 expression. The higher ratio in Common DC progenitor and classical type 1 DCs suggests that these APCs are expressing higher amounts of MR1 at the surface as opposed to intracellularly (Figure 4C). Overall, we show that MR1+ APCs are present at the maternal fetal interface, suggesting a path for MR1-dependent activation of MAIT cells. Coincidentally, we found no difference between surface/intracellular expression of MR1 between decidua basalis and parietalis APCs.
3.5. Activated MAIT cells in Decidua Basalis upregulate IFNγ and TNFα rather than Granzyme B
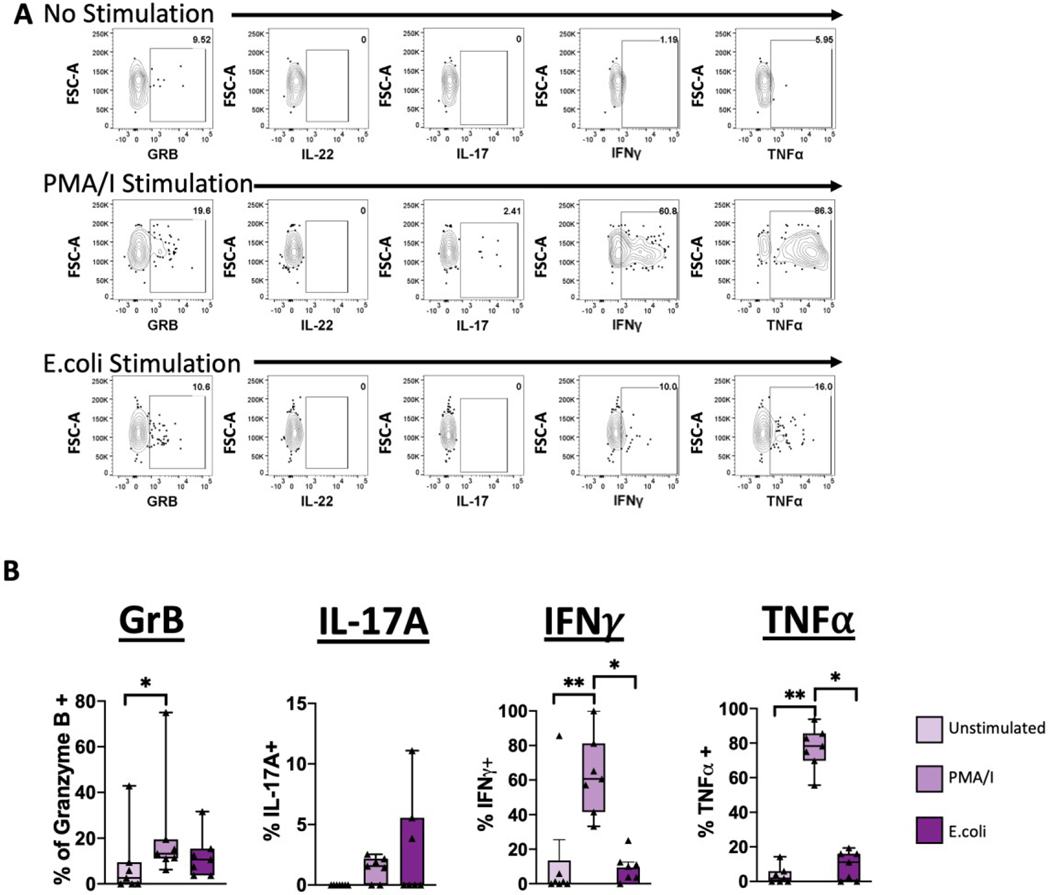
MAIT cells are able to produce a wide range of cytokines which is dependent on the nature of the stimulus (54). In previous studies, production of IFNγ, Granzyme B, and perforin have been observed in MAIT cells from the maternal-fetal interface (3). Based on these observations, we asked whether production of cytokines was consistent in term human decidua (Figure 5).
Figure 5. MAIT cells skew towards IFNγ and TNFα production.
Decidual MAIT cells were activated with PMA/Ionomycin or E.coli and production of cytokines was assessed. (A) Representative gating scheme of MAIT cells after activation showing the production of Granzyme B (GRB), IL-22, IL-17A, IFNγ and TNFα. (B) Frequency of GRB, IL-17A, IFNγ, and TNFα positive MAIT cells after activation. Decidua basalis, n = 6. Data represented as max/min, median, and 25 and 75th percentiles. Statistical significance was determined by One-way ANOVA, followed by Tukey post-hoc tests. *p < 0.05, **p < 0.005.
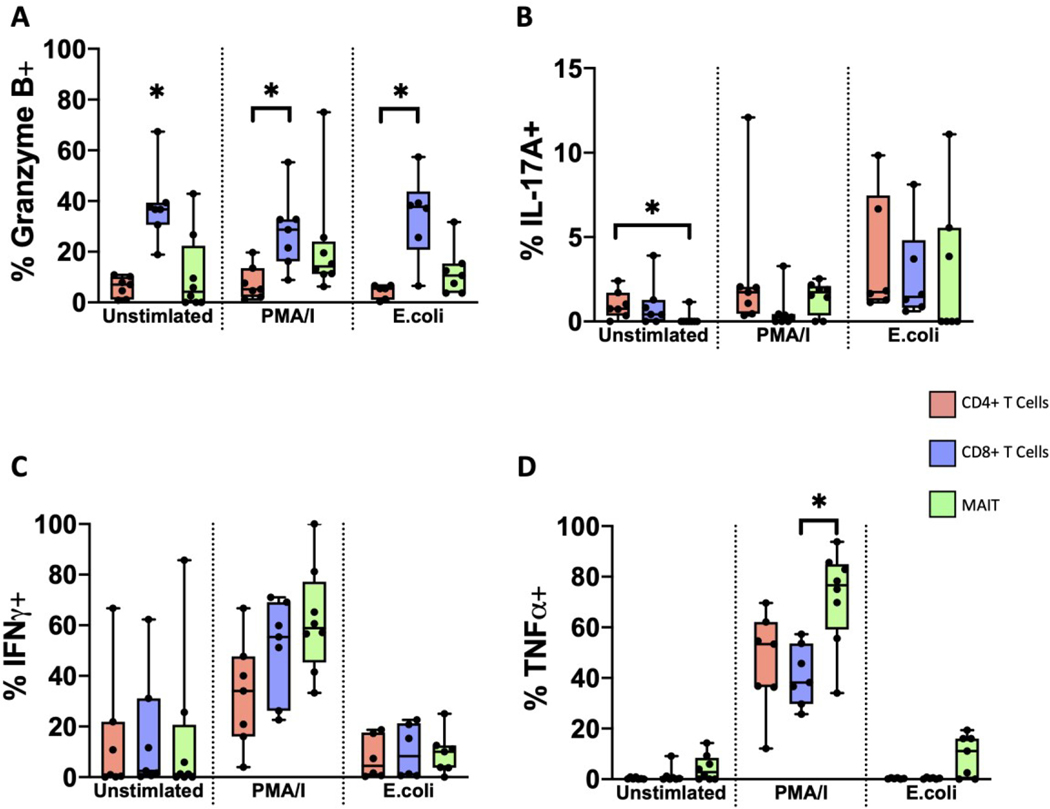
Granzyme B is a cytotoxic molecule produced by CD8+ and natural killer cells (55,56). MAIT cells from various tissues also have the capacity of producing Granzyme B, as part of their effector function (10,11). After activation with PMA/Ionomycin, we found that MAIT cells from term human decidua basalis had a slightly greater proportion of Granzyme B+ cells, compared to unstimulated cells (Figure 5B). Interestingly, we found no difference in Granzyme B production in E.coli activated MAIT cells, suggesting that there is a capacity for Granzyme B production but only with the appropriate stimulus. Interestingly, the proportion of Granzyme B+ cells was higher in decidual CD8+ T cells, compared to both CD4+ T cells and MAIT cells (Figure 6A), implying that CD8+ T cells but not MAIT cells might be the main producers of Granzyme B at the maternal-fetal interface.
Figure 6. MAIT cells display a different functional profile compared to conventional T cells.
Frequency of (A) GRB (B) IL-17A, (C) IFNγ, and (D) TNFα positive CD4+, CD8+, and MAIT cells after activation. Decidua basalis, n = 6. Data represented as max/min, median, and 25 and 75th percentiles. Statistical significance was determined by One-way ANOVA, followed by Tukey post-hoc tests, within treatment group. *p < 0.05.
MAIT cells have the capacity to secrete TH17-type cytokines, IL-22 and IL17A (10,15,20) in appropriate contexts. We therefore asked if MAIT cells from term decidua basalis have the capacity of producing IL-22 and IL-17A (Figure 5). Surprisingly, we found that IL-22 was not produced in an appreciable level by decidual MAIT cells under any of the treatments (Figure 5A). However, we found that the proportion of IL-17A+ MAIT cells trended higher in both the PMA/Ionomycin and E.coli stimulated groups, although this did not reach significance (Figure 5B). Moreover, we did find that CD4+ T cells had a higher proportion of IL-17A+ cells at baseline, compared to both CD8+ T cells and MAIT cells (Figure 6B).
IFNγ, an important cytokine often elicited in response to infections and tissue damage (57) and involved in decidualization (58,59), is primarily produced by TH1, CD8+ T cells and natural killer cells (57,60). Because MAIT cells in various tissues have also been shown to produce IFNγ (61–63), we assessed the production of IFNγ in decidual MAIT cells (Figure 5). The proportion of IFNγ+ MAIT cells was higher in those activated with PMA/Ionomycin (Figure 5B). We did not observe significant differences between the proportion of IFNγ+ conventional T cells and MAIT cells across all treatments (Figure 6C), suggesting a level of redundancy in induced IFNγ production at the maternal-fetal interface, or strict transcriptional control of IFNγ levels locally.
Lastly, we asked whether decidual MAIT cells have the capacity of producing TNFα, an important cytokine involved in the pro-inflammatory response (64). Similar to IFNγ, we found that the proportion of TNFα+ MAIT cells was highest under PMA/Ionomycin activation (Figure 5B). Furthermore, we found that when compared to conventional T cells, MAIT cells maintained a higher proportion of TNFα+ cells (Figure 6D). Overall, our results indicate the preferential production of IFNγ and TNFα by MAIT cells, while maintaining the capacity of Granzyme B production, albeit at lower levels than conventional T cells.
4. Discussion
The maternal-fetal interface is a unique mucosal site wherein balance between tolerance and protection is pivotal for a successful pregnancy. Imbalance in this system has been tied to pregnancy complications such as preeclampsia and preterm birth. Although restricted in its immune composition, the decidua maintains a unique immune cell population consisting primarily of decidual NK cells (65) but that also maintains a distinct distribution of T cell subsets (24,66,67). Recently, the innate-like T cell subset MAIT cells, important players in the maintenance of tissue homeostasis (51,62), have been identified in both the endometrium and the decidua (3,68). Here, we provide additional confirmation for the presence of MAIT cells in term human decidua and demonstrate, with the use of novel scRNA-seq technologies (CITE-seq), that decidual and peripheral MAIT cells maintain different gene expression profiles. Furthermore, our analysis of decidual APCs show the expression of the antigen-presenting molecule MR1, known restricting molecule to the MAIT TCR. Lastly, we show that decidual MAIT cells are skewed towards IFNγ and TNFα production, with the use of novel scRNA-seq technologies (CITE-seq),
We first confirmed the presence of MAIT cells in human decidua (Figures 1 and 2), in line with recent studies identifying MAIT cells in the intervillous space and the decidua (3–5). Interestingly, the abundance of decidual MAIT cells observed by us and others (3–5) is similar to that seen in the endometrium (15,52), suggesting that pregnancy does not alter the frequency of MAIT cells in the mucosa of female reproductive organs. Decidual MAIT cells identified in this study were also primarily CD8-positive, similar to other studies conducted in reproductive (3,15,52) and other tissues (69–71). It has also been reported that decidual parietalis MAIT cells are more activated than their decidua basalis and intervillous blood counterparts based on the expression of CD69, HLA-DR, CD38, and PD-1 (3,4). Even though we did not assess the expression of these proteins by flow cytometry, our scRNA-seq data support these previous observations, with decidual basalis MAIT cells maintaining a global gene expression profile indicative of attenuation (Figure 2D). Interestingly, CD69 is also a marker of tissue-residency (72), which was upregulated in decidua basalis MAIT cells compared to PBMCs (Figure 2). This hints to a possible division of labor between decidua basalis and parietalis MAIT cells, that is influenced by location, one being adjacent to the placenta and other to fetal membranes. Indeed, we found that Ccl4, a trophoblast chemoattractant (73), was, in fact, upregulated in decidua basalis MAIT cells (Figure 2), partly supporting our hypothesis. However, due to limitations in cellular recovery, we were not able to sequence full RNA transcriptomes from individual MAIT cells from the decidua parietalis to perform a direct comparison.
MAIT cells share many features with other tissue-resident lymphocytes (11,71,72). It has been suggested that at the maternal-fetal interface, MAIT cells might be recruited from the periphery by the chemoattractant MIF secreted by the placenta (5). This is further evidenced by a lower frequency of MAIT cells in the peripheral blood of pregnant individuals compared to non-pregnant, suggesting that MAIT cells are recruited from the periphery (3,5) to the intervillous space. Studies in transplanted uteri have also shown that endometrial MAIT cells are replaced by recipient MAIT cells (52), suggesting that MAIT cells residing in the endometrium are recruited from the periphery where they then obtain tissue-resident characteristics. Our data similarly support the idea that decidual MAIT cells obtain tissue-residency once recruited from the periphery. Specifically, the decidual MAIT cells we isolated lack expression of CD103, one of the hallmark integrins indicative of tissue residency (52,71,72), however, they maintain a specific gene expression signature compared to matched peripheral MAIT cells (Figure 2). These observations support recruitment of MAIT cells from the periphery into the decidual environment and that decidual MAIT cells are in the process of establishing themselves as decidual residents. One cannot discount the possibility, however, that decidual MAIT cells are composed of two populations: a tissue-resident and a transient population.
Functionally, MAIT cells have been recognized for their ability to respond rapidly to microbes at mucosal sites (6,74,75). MAIT cells also have the capacity of responding to non-antigenic triggers, such as cytokines (61). The presence of MAIT cells in the decidua opens a host of questions regarding their role in pregnancy. In the endometrium, MAIT cells have been shown to skew towards an IL-17A-producing phenotype after E.coli stimulation (15). However, in the decidua, MAIT cells consistently skew towards IFNγ production under microbial stimulation (4,52) a finding supported by our results as well (Figure 5). This suggests that MAIT cells might play an analogous role to that of dNKs at the maternal-fetal interface (58,73), where IFNγ production leads to angiogenesis and decidual remodeling, rather than clearance of microbial populations. Similar tissue-repair roles of MAIT cells have been uncovered in other tissues (62,76,77), coincidentally all requiring the presence of microbiota. The expression of the MR1 molecule by decidual APCs (3) (Figure 4), suggests that decidual MAIT cells can be activated by local decidual microbes. However, the recent debate on whether the placenta maintains a local microbial community (78–81), adds a layer of complexity to our understanding of MAIT cells in the decidua, as all evidence thus far indicates that the presence of microbes is necessary to induce tissue-repair functions in MAIT cells (62,76,77).
Single cell transcriptomics has emerged as a powerful tool to understand gene regulation mechanisms in various tissues, including tissues of the maternal-fetal interface (82–85). To the best of our knowledge, this is the first study to identify decidual MAIT cells using scRNA-seq, as others have identified them only in the blood (83,84) using this technology. This discrepancy can be attributed to experimental design, as we targeted only CD3+ T cells for sequencing while the other two studies (83,84) had a broader cellular target. Use of CITE-seq confirmed MAIT cell phenotype of T cells in the decidua based on surface expression of TCR-vα7.2 and CD161. Our single cell data show that decidual MAIT cells are programmed for attenuated responsiveness compared to peripheral MAIT cells, which is in agreement with a recent study showing a pro-inflammatory profile in peripheral MAIT cells from patients impacted by recurrent pregnancy loss (84). Further studies analyzing matched decidual and peripheral samples from disease states should shed light on specific MAIT cell programming.
Lastly, comparisons of lineage-defining transcription factor expression found that overall MAIT cells isolated from the decidua are different from non-pregnant, non-matched MAIT cells, while those isolated from pregnant individuals (matched decidua and peripheral) are the most similar (Figure S4 and S5). This suggests that not only local tissue development (of the decidua) but also global (hormones) changes induced by pregnancy have a specific transcriptional effect on MAIT cells. This observation is limited as different methods (flow cytometry, scRNA-seq) were used to compare pregnant vs. non-pregnant. Moreoever, analysis of MAIT cells pre- and post-pregnancy would be needed to highlight transcriptional changes due to pregnancy. Nonetheless, our results emphasize the importance of considering transcriptional changes, independent of phenotypic characteristics, incurred by immune cells in the context of pregnancy.
Supplementary Material
Acknowledgements:
We thank the NIH tetramer facility at Emory University for graciously providing the tetramers. The Flow Cytometry Core for expert assistance. Jenny E. Gumperz, Nick Zumwalde for providing E.coli samples and their expertise. JV was supported by NIH Ruth I. Kirschtein National Research Award (T32-HD041921), NIH TEAM-Science (R25 GM083252), UW SciMed GRS Fellowship. M.C. was supported by WISE Summer Research Grant. AKS was supported by grant K12HD000849–28 awarded to the Reproductive Scientist Development Program by the Eunice Kennedy Shriver National Institute of Child Health & Human Development, March of Dimes Basil O’Connor Award (5-FY18–541) and Burroughs Wellcome Fund Preterm Birth Award (1019835). Additional research support (to AKS) provided by American Society for Reproductive Medicine, March of Dimes, and Burroughs Wellcome Fund, as part of the Reproductive Scientist Development Program Supplement and Seed Programs. IMO acknowledges support by the Clinical and Translational Science Award (CTSA) program (ncats.nih.gov/ctsa), through the National Institutes of Health National Center for Advancing Translational Sciences (NCATS), grants UL1TR002373 and KL2TR002374. Additional research support (to IMO) provided by the Career Enhancement
Program award from the Specialized Program of Research Excellence (SPORE) program through the NIH National Institutes of Health for Dental and Craniofacial Research (NIDCR) and National Cancer Institute (NCI) grant P50DE026787, COVID-19 Supplement from the National Institutes of Health grant 2U19AI104317–06 (to IMO via James Gern), the Hartwell Foundation, and the Wisconsin Partnership Program. UWCCC Cancer Center Support Grant (CCSG) for use Share Resources BD LSR Fortessa (NIH Grant No. 1S00OD018202–01).
Footnotes
Conflict of interest disclosure statement. All of the co-authors declare that they do not have any relationships that could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to the manuscript being submitted for review.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/aji.13495.
References
- 1.Kanellopoulos-Langevin C, Caucheteux SM, Verbeke P, Ojcius DM. Tolerance of the fetus by the maternal immune system: role of inflammatory mediators at the feto-maternal interface. Reprod Biol Endocrinol. 2003;1(121). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.PrabhuDas M, Bonney E, Caron K, Dey S, Erlebacher A, Fazleabas A, et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat Immunol. 2015. April;16(4):328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solders M, Gorchs L, Erkers T, Lundell A-C, Nava S, Gidlöf S, et al. MAIT cells accumulate in placental intervillous space and display a highly cytotoxic phenotype upon bacterial stimulation. Sci Rep. 2017. December;7(1):6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solders M, Gorchs L, Gidlöf S, Tiblad E, Lundell A-C, Kaipe H. Maternal Adaptive Immune Cells in Decidua Parietalis Display a More Activated and Coinhibitory Phenotype Compared to Decidua Basalis. Stem Cells Int. 2017;2017:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solders M, Kaipe H. Recruitment of MAIT Cells to the Intervillous Space of the Placenta by Placenta-Derived Chemokines. Front Immunol. 2019;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koay H-F, Godfrey DI, Pellicci DG. Development of mucosal-associated invariant T cells. Immunol Cell Biol. 2018. July;96(6):598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003. March;422(6928):164–9. [DOI] [PubMed] [Google Scholar]
- 8.Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013. October 21;210(11):2305–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori L, Lepore M, De Libero G. The Immunology of CD1- and MR1-Restricted T Cells. Annu Rev Immunol. 2016. May 20;34(1):479–510. [DOI] [PubMed] [Google Scholar]
- 10.Keller AN, Corbett AJ, Wubben JM, McCluskey J, Rossjohn J. MAIT cells and MR1-antigen recognition. Curr Opin Immunol. 2017. June;46:66–74. [DOI] [PubMed] [Google Scholar]
- 11.Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 2011. January 27;117(4):1250–9. [DOI] [PubMed] [Google Scholar]
- 12.Jo J, Tan AT, Ussher JE, Sandalova E, Tang X-Z, Tan-Garcia A, et al. Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver. Walker CM, editor. PLoS Pathog. 2014. June 26;10(6):e1004210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human Mucosal Associated Invariant T Cells Detect Bacterially Infected Cells. Marrack P, editor. PLoS Biol. 2010. June 29;8(6):e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuyama H, Isshiki T, Chiba A, Yamaguchi T, Murayama G, Akasaka Y, et al. Activation of mucosal-associated invariant T cells in the lungs of sarcoidosis patients. Sci Rep. 2019. December;9(1):13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017. January;10(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010. August;11(8):701–8. [DOI] [PubMed] [Google Scholar]
- 17.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014. May;5(1):3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howson LJ, Salio M, Cerundolo V. MR1-Restricted Mucosal-Associated Invariant T Cells and Their Activation during Infectious Diseases. Front Immunol. 2015. June 16;6(303). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat Commun. 2014. September;5(1):3866. [DOI] [PubMed] [Google Scholar]
- 20.Zumwalde NA, Haag JD, Gould MN, Gumperz JE. Mucosal associated invariant T cells from human breast ducts mediate a Th17-skewed response to bacterially exposed breast carcinoma cells. Breast Cancer Res. 2018. December;20(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gherardin NA, Souter MN, Koay H-F, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. 2018. May;96(5):507–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurioka A, Walker LJ, Klenerman P, Willberg CB. MAIT cells: new guardians of the liver. Clin Transl Immunol. 2016. August 19;5(8):e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Plazyo O, Romero R, Hassan SS, Gomez-Lopez N. Isolation of Leukocytes from the Human Maternal-fetal Interface. J Vis Exp. 2015. May 21;(99):52863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez J, Chavarria M, Li Y, Lopez GE, Stanic AK. Computational flow cytometry analysis reveals a unique immune signature of the human maternal-fetal interface. Am J Reprod Immunol. 2018. January;79(1):e12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez J, Chasman DA, Lopez GE, Tyler CT, Ong IM, Stanic AK. Transcriptional and Functional Programming of Decidual Innate Lymphoid Cells. Front Immunol. 2020. January 24;10:3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias J, Sandberg JK, Leeansyah E. Extensive Phenotypic Analysis, Transcription Factor Profiling, and Effector Cytokine Production of Human MAIT Cells by Flow Cytometry. In: Lugli E, editor. T-Cell Differentiation. New York, NY: Springer New York; 2017. p. 241–56. (Methods in Molecular Biology; vol. 1514). [DOI] [PubMed] [Google Scholar]
- 27.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016. November;539(7628):309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019. June;177(7):1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017. September;14(9):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, et al. CD161 Defines a Transcriptional and Functional Phenotype across Distinct Human T Cell Lineages. Cell Rep. 2014. November;9(3):1075–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise Development of MAIT Cells in Mouse and Human. Cerundolo V, editor. PLoS Biol. 2009. March 10;7(3):e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore MJ, Blachere NE, Fak JJ, Park CY, Sawicka K, Parveen S, et al. ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. eLife. 2018. May 31;7:e33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das T, Chen Z, Hendriks RW, Kool M. A20/Tumor Necrosis Factor α-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models. Front Immunol. 2018. February 21;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu D, Liu L, Ji X, Gao Y, Chen X, Liu Y, et al. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat Immunol. 2015. December;16(12):1263–73. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Xia L, Chen J, Zhang S, Martin V, Li Q, et al. Stress–glucocorticoid–TSC22D3 axis compromises therapy-induced antitumor immunity. Nat Med. 2019. September;25(9):1428–41. [DOI] [PubMed] [Google Scholar]
- 36.Hwang SS, Lim J, Yu Z, Kong P, Sefik E, Xu H, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020. March 13;367(6483):1255–60. [DOI] [PubMed] [Google Scholar]
- 37.Sekiya T, Kashiwagi I, Inoue N, Morita R, Hori S, Waldmann H, et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011. September;2(1):269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebmann M, Hucke S, Koch K, Eschborn M, Ghelman J, Chasan AI, et al. Nur77 serves as a molecular brake of the metabolic switch during T cell activation to restrict autoimmunity. Proc Natl Acad Sci. 2018. August 21;115(34):E8017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tegla CA, Cudrici CD, Nguyen V, Danoff J, Kruszewski AM, Boodhoo D, et al. RGC-32 is a novel regulator of the T-lymphocyte cell cycle. Exp Mol Pathol. 2015. June;98(3):328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenkel JM, Masopust D. Tissue-Resident Memory T Cells. Immunity. 2014. December;41(6):886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017. September;20(12):2921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannan NJ, Jones RL, White CA, Salamonsen LA. The Chemokines, CX3CL1, CCL14, and CCL4, Promote Human Trophoblast Migration at the Feto-Maternal Interface1. Biol Reprod. 2006. May 1;74(5):896–904. [DOI] [PubMed] [Google Scholar]
- 43.Filen S, Lahesmaa R. GIMAP Proteins in T-Lymphocytes. J Signal Transduct. 2010;2010:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muri J, Heer S, Matsushita M, Pohlmeier L, Tortola L, Fuhrer T, et al. The thioredoxin-1 system is essential for fueling DNA synthesis during T-cell metabolic reprogramming and proliferation. Nat Commun. 2018. December;9(1):1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simula L, Pacella I, Colamatteo A, Procaccini C, Cancila V, Bordi M, et al. Drp1 Controls Effective T Cell Immune-Surveillance by Regulating T Cell Migration, Proliferation, and cMyc-Dependent Metabolic Reprogramming. Cell Rep. 2018. December;25(11):3059–3073.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011. April;23(2):220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eberl G. RORγt, a multitask nuclear receptor at mucosal surfaces. Mucosal Immunol. 2017. January;10(1):27–34. [DOI] [PubMed] [Google Scholar]
- 48.Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-Bet and Eomes in Peripheral Human Immune Cells. Front Immunol. 2014. May;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonetta F, Pradier A, Roosnek E. T-bet and Eomesodermin in NK Cell Development, Maturation, and Function. Front Immunol. 2016. June 20;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohamed R, Lord GM. T-bet as a key regulator of mucosal immunity. Immunology. 2016. April;147(4):367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaipe H, Raffetseder J, Ernerudh J, Solders M, Tiblad E. MAIT Cells at the Fetal-Maternal Interface During Pregnancy. Front Immunol. 2020. August 19;11:1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bister J, Crona Guterstam Y, Strunz B, Dumitrescu B, Haij Bhattarai K, Özenci V, et al. Human endometrial MAIT cells are transiently tissue resident and respond to Neisseria gonorrhoeae. Mucosal Immunol. 2020. August 5; [DOI] [PubMed] [Google Scholar]
- 53.Karamooz E, Harriff MJ, Lewinsohn DM. MR1-dependent antigen presentation. Semin Cell Dev Biol. 2018. December;84:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Godfrey DI, Koay H-F, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019. September;20(9):1110–28. [DOI] [PubMed] [Google Scholar]
- 55.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015. June;15(6):388–400. [DOI] [PubMed] [Google Scholar]
- 56.Behr FM, Chuwonpad A, Stark R, van Gisbergen KPJM. Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells. Front Immunol. 2018. July 30;9:1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018. September;18(9):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashkar AA, Croy BA. Functions of uterine natural killer cells are mediated by interferon gamma production during murine pregnancy. Semin Immunol. 2001. August;13(4):235–41. [DOI] [PubMed] [Google Scholar]
- 59.Ashkar AA, Croy BA. Interferon-␥ Contributes to the Normalcy of Murine Pregnancy. 1999;61:493–502. [DOI] [PubMed] [Google Scholar]
- 60.Mühl H, Pfeilschifter J. Anti-inflammatory properties of pro-inflammatory interferon-γ. Int Immunopharmacol. 2003. September;3(9):1247–55. [DOI] [PubMed] [Google Scholar]
- 61.Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or Cytokine-Activated CD8+ Mucosal-Associated Invariant T Cells Are Rapid Polyfunctional Effectors That Can Coordinate Immune Responses. Cell Rep. 2019. September;28(12):3061–3076.e5. [DOI] [PubMed] [Google Scholar]
- 62.Leng T, Akther HD, Hackstein C-P, Powell K, King T, Friedrich M, et al. TCR and Inflammatory Signals Tune Human MAIT Cells to Exert Specific Tissue Repair and Effector Functions. Cell Rep. 2019. September;28(12):3077–3091.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. 2019. October;19(10):643–57. [DOI] [PubMed] [Google Scholar]
- 64.Sedger LM, McDermott MF. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants – past, present and future. Cytokine Growth Factor Rev. 2014. August;25(4):453–72. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J, Dunk C, Croy AB, Lye SJ. To serve and to protect: the role of decidual innate immune cells on human pregnancy. Cell Tissue Res. 2016. January;363(1):249–65. [DOI] [PubMed] [Google Scholar]
- 66.Dimova T, Nagaeva O, Stenqvist A-C, Hedlund M, Kjellberg L, Strand M, et al. Maternal Foxp3 Expressing CD4+ CD25+ and CD4+ CD25− Regulatory T-Cell Populations are Enriched in Human Early Normal Pregnancy Decidua: A Phenotypic Study of Paired Decidual and Peripheral Blood Samples: TREG CELLS IN HUMAN PREGNANCY. Am J Reprod Immunol. 2011. July;66:44–56. [DOI] [PubMed] [Google Scholar]
- 67.Miller D, Gershater M, Slutsky R, Romero R, Gomez-Lopez N. Maternal and fetal T cells in term pregnancy and preterm labor. Cell Mol Immunol. 2020. May 28; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Human endometrial MAIT cells are transiently tissue resident and respond to Neisseria gonorrhoeae. Mucosal Immunol. 2020; [DOI] [PubMed] [Google Scholar]
- 69.Voillet V, Buggert M, Slichter CK, Berkson JD, Mair F, Addison MM, et al. Human MAIT cells exit peripheral tissues and recirculate via lymph in steady state conditions. JCI Insight. 2018. April 5;3(7):e98487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019. January 7;216(1):133–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgård J, Dias J, et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur J Immunol. 2019. January;49(1):133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan X, Rudensky AY. Hallmarks of Tissue-Resident Lymphocytes. Cell. 2016. March;164(6):1198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006. September;12(9):1065–74. [DOI] [PubMed] [Google Scholar]
- 74.Berkson JD, Prlic M. The MAIT conundrum – how human MAIT cells distinguish bacterial colonization from infection in mucosal barrier tissues. Immunol Lett. 2017. December;192:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ussher JE, Klenerman P, Willberg CB. Mucosal-Associated Invariant T-Cells: New Players in Anti-Bacterial Immunity. Front Immunol. 2014. October 8;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han S-J, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019. October 25;366(6464):eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and In Vivo Evolution of the MAIT Cell Transcriptome in Mice and Humans Reveals Tissue Repair Functionality. Cell Rep. 2019. September;28(12):3249–3262.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med. 2014. May 21;6(237):237ra65–237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bushman FD. De-Discovery of the Placenta Microbiome. Am J Obstet Gynecol. 2019. March;220(3):213–4. [DOI] [PubMed] [Google Scholar]
- 80.Leiby JS, McCormick K, Sherrill-Mix S, Clarke EL, Kessler LR, Taylor LJ, et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018. December;6(1):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol. 2016. May;214(5):627.e1–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, et al. A single-cell survey of the human first-trimester placenta and decidua. Sci Adv. 2018. October;4(10):eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018. November;563(7731):347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang F, Jia W, Fan M, Shao X, Li Z, Liu Y, et al. Single-cell Immune Landscape of Human Recurrent Miscarriage. Genomics Proteomics Bioinformatics. 2021. January;S1672022921000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo C, Cai P, Jin L, Sha Q, Yu Q, Zhang W, et al. Single-cell profiling of the human decidual immune microenvironment in patients with recurrent pregnancy loss. Cell Discov. 2021. December;7(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.