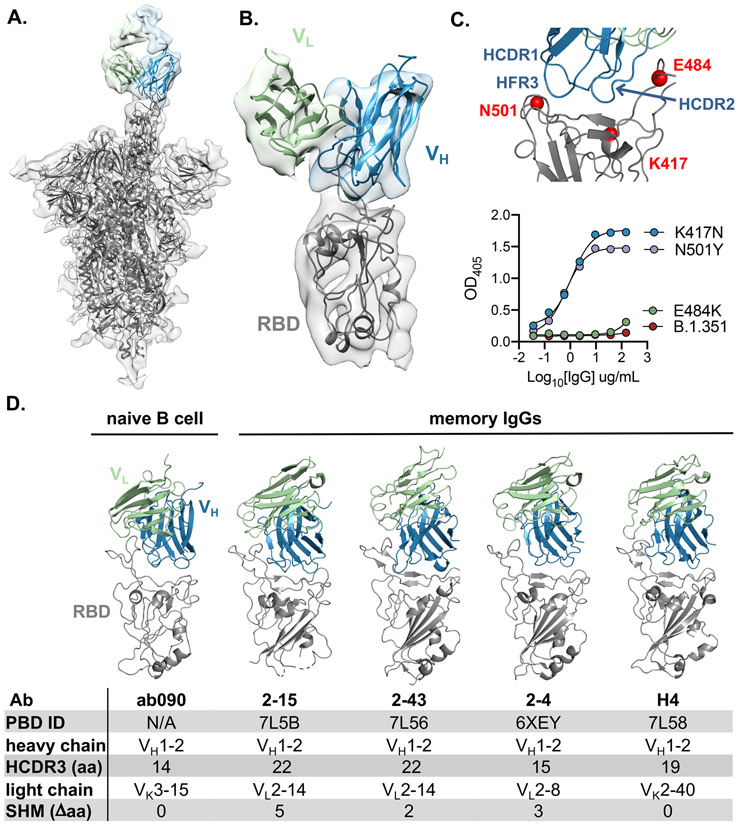
Fig. 4. ab090 recognizes the SARS-CoV-2 RBM.
(A) Cryo-EM structure of the SARS-CoV-2 spike trimer (grey) with ab090 Fab bound to one RBD in the up position. (B) ab090 recognizes the SARS-CoV-2 RBM with through a paratope centered on the VH (blue). (C) Close-up view showing the approximate locations of HCDR loops proximal to the RBM epitope and B.1.351 RBD mutations highlighted in red (top). ELISA binding reactivity of ab090 to individual mutations from B.1.351 RBD (bottom). (D) ab090 and representative IGHV1-2 neutralizing antibodies isolated from memory B cells from convalescent COVID-19 donors bound to SARS-CoV-2 RBD (grey). Structures are shown in the same relative orientation in each panel with PBD codes and sequence attributes listed in the below.