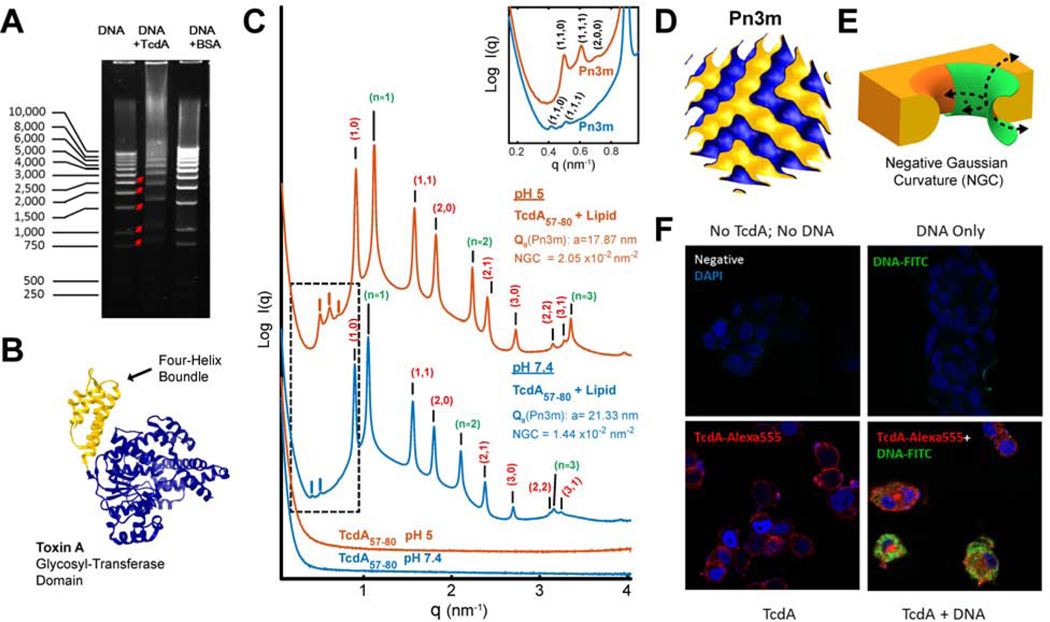
Figure 1. C. difficile TcdA binds DNA and facilitates DNA’s cellular entry (A-E).
A) TcdA binds to DNA. Gel electrophoresis showing that the incubation of TcdA (10μg) with a 1kb DNA ladder (2μg) retarded DNA mobility. Incubation of DNA with BSA (10μg) acted as a negative control.
B) Four-helix bundle in the glucosyltransferase domain GTD. Solved 3D crystal structure of the N-terminal glucosyltransferase domain from TcdA (PDB ID: 4DMV), highlighting the position of the membrane targeting N-terminal four-helix bundle motif (yellow).
C) TcdA four-helix bundle segment, TcdA57–80, generates NGC necessary for membrane permeabilization. SAXS spectra from 20/70/10 PS/PE/Chl model membranes incubated with TcdA57–80, a 24 amino acids fragment in the TcdA N-terminal four-helix bundle, at a 1:4 peptide-to-lipid charge ratio. At both pH 7.4 and pH 5.0, TcdA57–80 induces NGC in the form of a Pn3m QII cubic phase (insert plot). The additional observed reflections for the positive curvature hexagonal (red indices) and lamellar (green indices) phases have been assigned on the curves, with periodicity of 8.0 and 5.7nm at pH 5, and 8.1 and 6.0nm at pH 7.4, respectively. No Bragg reflections were observed for the peptide solutions alone (bottom). Only hexagonal phases were for the membrane SUVs suspensions at both pHs (Supplementary Figure S1). To facilitate visualization, spectra have been manually offset vertically by a multiplicative factor.
D) Reconstruction of a Pn3m surface. 3D reconstruction depicting a Pn3m QII cubic phase, with continuous surfaces forming NGC at every point.
E) Illustration of NGC geometry. 3D rendering of the saddle-splay inherent to negative Gaussian curvature geometry (green) as superimposed on a mechanism of membrane remodeling.
F) TcdA facilitates cellular uptake of DNA. HT29 colonic epithelial cells were treated with medium (negative) or DNA-FITC (1μg/mL, green) alone or along with Alexa 555 labeled TcdA (100nM, red) for 2 hours at 37°C. Cells were fixed with Cyto-Fix at 4°C for 15 minutes and nuclei were stained with DAPI (blue). Significantly increased plasmid DNA was observed in the cytosol of TcdA-treated colonocytes (bottom right panel).