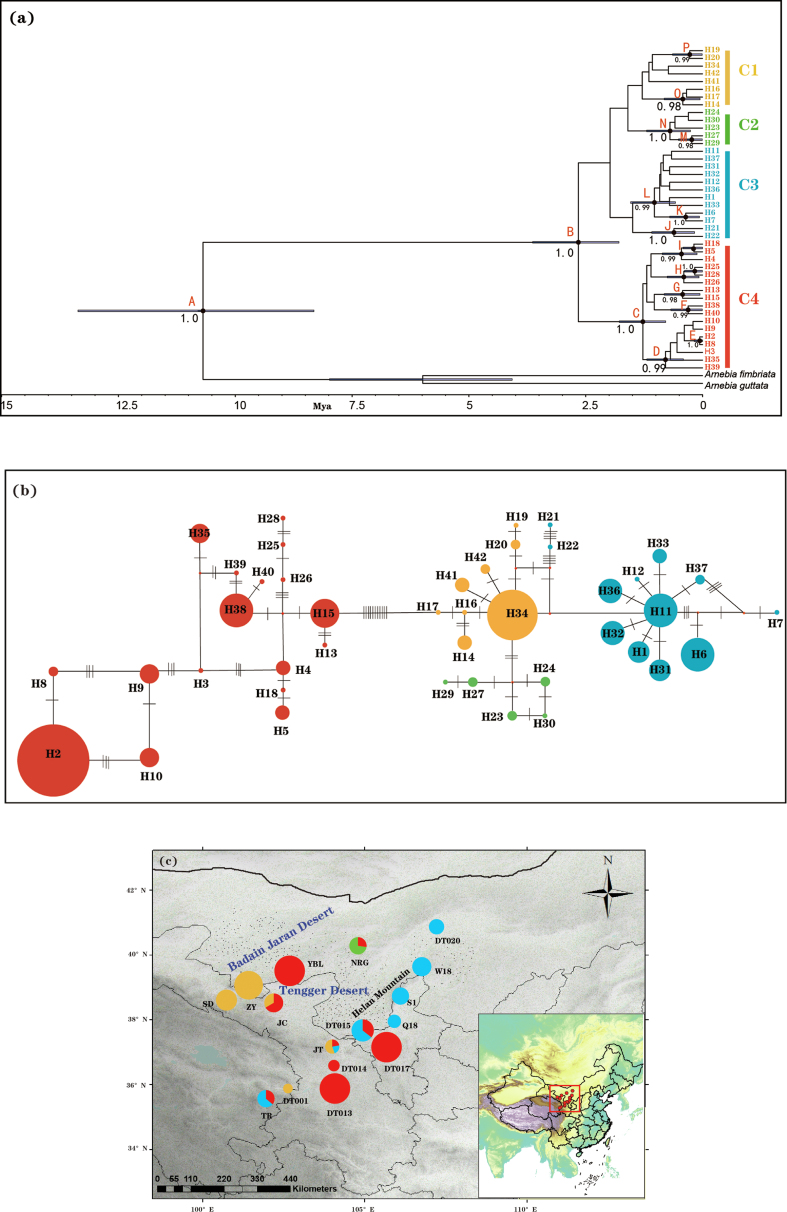
Fig. 4.
(a) BEAST-derived chronogram of divergence times within Arnebia szechenyi based on three chloroplast DNA fragments (psbA-trnH, psbB-psbF, trnL-trnF). Blue bars on each node indicate 95% highest posterior density (HPD) intervals for the TMRCA estimates. Posterior probabilities (PP) > 0.95 are shown above nodes. (b) The most parsimonious network of the haplotypes. Circles sizes are proportional to chlorotype frequencies within the total sample. Small red dots represent hypothetical unsampled or extinct ancestral haplotypes, and lines between circles represent single mutational steps. Pie colors correspond the different clades identified. (c) Geographical distribution of the four cpDNA clades. Pie charts show the relative proportion of each cpDNA clade within each population.