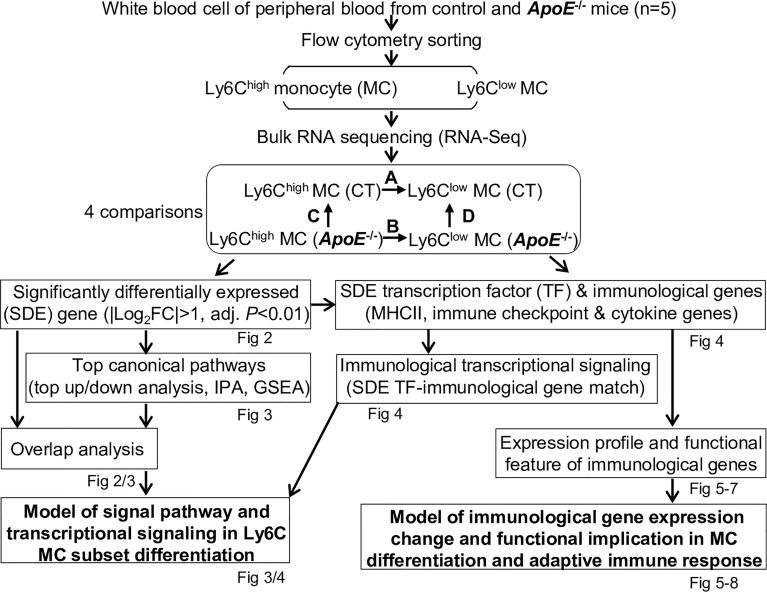
Figure 1.
Overall strategy of the identification of molecular signaling in Ly6C MC subset differentiation and adaptive immune response in control and ApoE -/- mice. RNA-Seq were performed in Ly6Chigh (CD11b+Ly6G−Ly6Chigh) and Ly6Clow (CD11b+Ly6G−Ly6Clow) MC isolated by flow cytometry sorting from peripheral blood of C57/BL6 control and ApoE -/- mice. Transcriptome data were analyzed by performing four pairs of comparisons: (A) Ly6Chigh vs Ly6Clow (CT), (B) Ly6Chigh vs Ly6Clow (ApoE-/- ), (C) ApoE-/- vs CT (Ly6Chigh), (D) ApoE-/- vs CT (Ly6Clow). SDE genes were identified by using the criteria of |Log2FC| more than 1 (2-FC) and adjusted P value less than 0.01. Top canonical pathways were recognized by top-down analysis using IPA with |Z-score|>2, P value<0.05. Overlapped analysis were performed for canonical pathways between groups. SDE TF and three sets of SDE immunological genes (MHCII, immune checkpoint and cytokine genes) were identified. Immunological SDE genes were matched with corresponding upstream SDE TF by IPA upstream analysis. Models of signal pathway and transcriptional signaling of Ly6C MC subset differentiation were developed. Expression profile and functional feature of immunological SDE genes were characterized. Model of immunological gene expression change and functional implication in MC differentiation and adaptive immune response in MC subsets of both mice were developed. CT, control, ApoE, Apolipoprotein E; FC, fold change; RNA-seq, RNA-sequencing; MC, monocyte; SDE, significant differentially expressed; IPA, Ingenuity Pathway Analysis; GSEA, gene set enrichment analysis; TF, transcription factor.