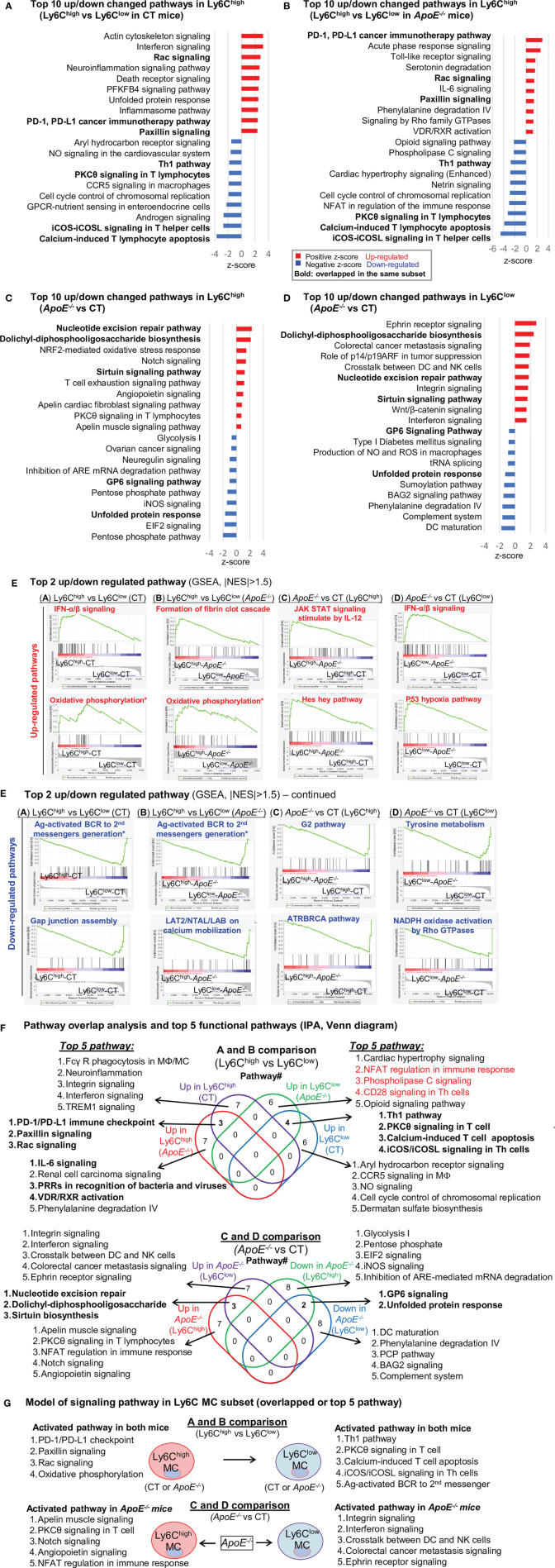
Figure 3.
Canonical pathway analysis for SDE genes in four comparison groups (Top 10 up/down changed pathways), (A). CT Ly6Chigh vs CT Ly6Clow; (B). ApoE-/- Ly6Chigh vs ApoE-/- Ly6Clow; (C). Ly6Chigh (ApoE-/- vs CT); (D). Ly6Clow (ApoE-/- vs CT). Top up/down changed canonical pathways were identified by using IPA software with the criteria of adjusted P value<0.05. Blue bar indicates a negative z-score and down-regulated pathways. Red bar indicates a positive z-score and up-regulated pathways. Bold letters indicate overlapped pathway in the same subset. (E) Top 2 up/down regulated pathway. The most enriched significant pathways from GSEA study with threshold of |NES|>1.5 are marked (red=up-regulated, blue=down-regulated). The green curve corresponds to the enriched score. The y-axis indicates the enriched score. The x-axis displaces genes (vertical black lines) represented in their pathway gene set. Rainbow bands represent the corresponding enriched score of the genes (red for positive and blue for negative correlation). The top 20 upregulated or downregulated GSEA pathways were shown in Supplementary Table 1 . (F) Pathway overlap analysis and top 5 functional pathways. Venn diagram summarized the overlap of top 10 pathway presented in (A–D) and listed the top 5 pathways in four pairs of comparisons. (G) Model of signaling pathway in Ly6C MC subset (overlapped or top 5 pathway). Four activated pathways in Ly6Chigh MC and 5 activated pathway in Ly6Clow MC were overlapped in A and B comparison. ApoE -/- induced top 5 activated pathway in both Ly6Chigh MC and Ly6Clow MC. MC, monocyte; MΦ, macrophage; TREM1, The triggering receptor expressed on myeloid cells 1; GPCRs, G-protein-coupled receptors; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; SLE, Systemic Lupus Erythematosus, Th1, T helper 1; PKCθ, Protein Kinase C Theta; IL-7, Interleukin 7; NFAT, Nuclear factor of activated T-cells; CHK, Csk-homologous kinase; nNOS, neuronal nitric oxide synthase; PXR, pregnane X receptor; CAR, constitutive androstane receptor. NO, Nitric Oxide; ROS, Reactive Oxygen Species; PRRs, Pattern Recognition Receptors.