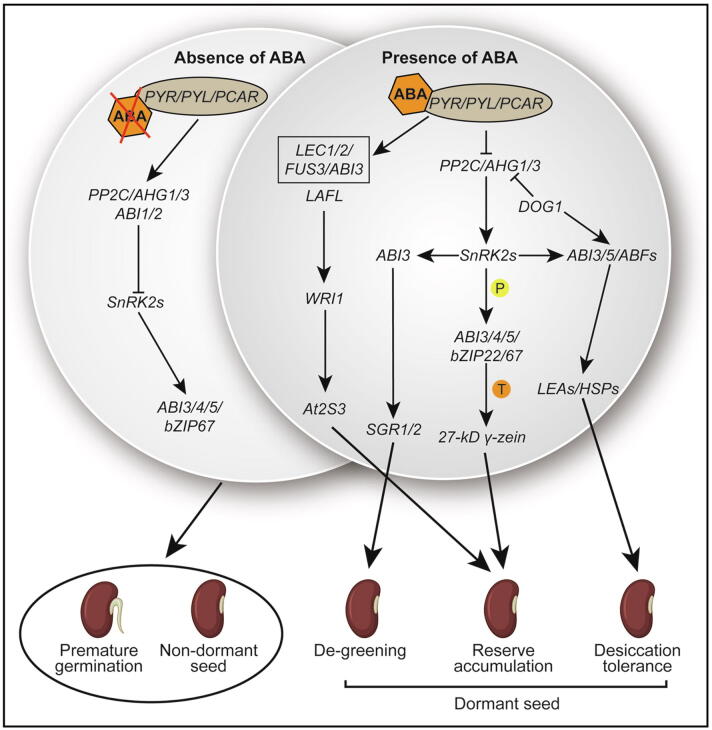
Fig. 2.
The ABA signaling pathway is involved in seed development. Left, in the absence of ABA: Receptors PYLs release and activate protein phosphatase 2C (PP2C) such as ABI1/2 and AGH1/3. Downstream SNF1-RELATED PROTEIN KINASE subfamily (SnRK2s) genes are inactivated by active PP2C which leads to premature germination and the non-dormant seed through repression of lots of transcription factors such as ABI1/2/3/4/5 and bZIP67. Right, in the presence of ABA: Receptors PYR/PYL/RCAR bind ABA and PP2C together to inhibit the activity of PP2C, which release the activity of SnRK2s and downstream transcription factors such as ABI3 by protein phosphorylation, then regulate downstream genes SGR1/2 function to mediate seed de-greening process. Additionally, the active LAFL (ABI3, FUS3, LEC1, and LEC2) network by ABA along with WRI1 regulates the At2S3 gene; an active bZIP22 function downstream of SnRK2s to promote gene transcription of 27-kD γ-zein for protein reserve accumulation in the seed. Along with seed de-greening and storage product accumulation, SnRK2s function upstream of ABI3/5 and ABFs to regulate LEAs and HSPs that are pivotal for desiccation tolerance. In other branches, DOG1 also plays a role upstream of ABI3/5/ABFs as well as functions as a repressor of PP2Cs (AHG1/3) to involve seed desiccation tolerance acquirement. In combination, all key ABA signaling components (SnRK2s, ABI3, ABI4, ABI5, ZmbZIP22, bZIP67, and ABFs) are involved in storage product accumulation, de-greening, and desiccation tolerance with different function pathways to provide a mature and dormant seed. Letters “P” and “T” in the color circles indicate the two manners of protein phosphorylation and gene transcription regulation, respectively. Activated and repressive effects are shown by arrows and bars, respectively.