Figure 1.
Single-cell RNA sequencing reveals immune and epithelial cell heterogeneity in paired normal-cancer samples
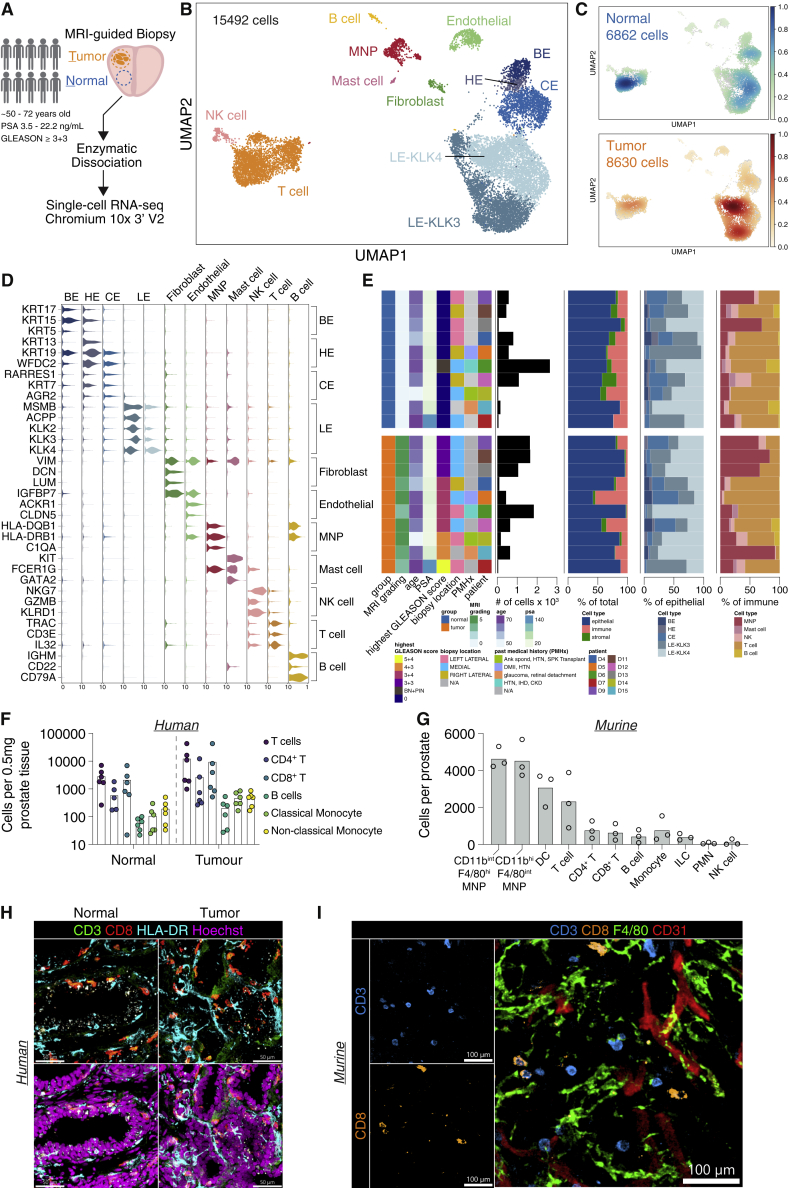
(A) Schematic describing experimental set-up for 10x genomic single-cell RNaseq of matched tumor-normal prostate samples from n = 10 patients.
(B) UMAP of 15,492 cells post-QC from all prostate samples.
(C) UMAP of embedding density of source of samples (normal—green to blue gradient, top; tumor—orange to red gradient, bottom).
(D) Violin plot of canonical marker genes for each cell types found in prostate. Gene expression values per cell are standardized to a range from 0 to 10.
(E) Patient demographics displayed as a color-coded heatmap and stacked bar charts of single-cell cell type proportions.
(F) Quantification of absolute cells counts by flow cytometry per 0.5mg of normal and malignant human prostatic tissue for the indicated immune subsets. Data are a combination of n = 6 donors with each dot representing an individual donor.
(G) Quantification of absolute cells counts by flow cytometry per murine prostate lobe for the indicated immune subsets. Each dot represents an individual mouse (n = 3 biological replicates). Abbreviations: MNP—mononuclear phagocyte; DC—dendritic cell; ILC—innate-like lymphoid cell; PMN—polymorphonuclear; NK natural killer (H) Confocal imaging of CD3, CD4 and CD8 in human normal and tumor prostate. Scale bars, 50 μm. (I) Confocal imaging of CD3, CD8, F4/80 and CD31 in normal murine prostate. Scale bars, 100 μm. See also Figure S1 and Table S1.