Figure 3.
A large fraction of rotation-selective neurons encode AHV
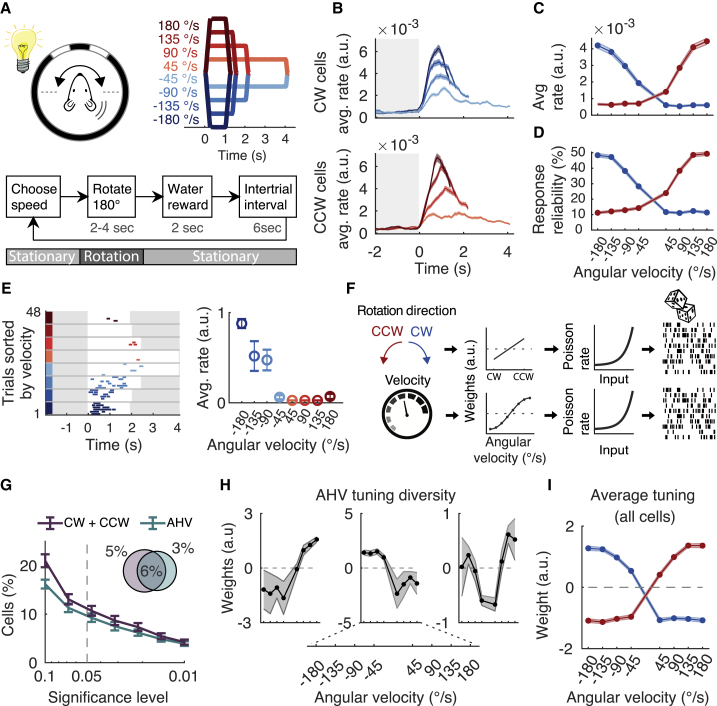
(A) (Top) Experimental configuration. Mouse is rotated 180° back and forth in CW and CCW directions and at different rotation speeds. The speed profiles are colored blue (CW) and red (CCW). (Bottom) Block diagram of the sequence of events for a single trial. The speed is randomly chosen for every trial.
(B) Response (average rate ± SEM) of all cells classified as CW (top) or CCW selective (bottom) and separated by rotation speed (4,928 cells, 10 mice, 14 FOVs).
(C) Response (average rate ± SEM) as a function of angular velocity, for CW (blue) and CCW (red) rotations. Same data as in (B). Only the first 1 s was analyzed, which is the duration of the fastest rotation.
(D) Response reliability (average ± SEM) as a function of angular velocity, for CW (blue) and CCW (red) rotations. Same data as in (B).
(E) (Left) Responses of an example CW cell for a whole session (deconvolved DF/F events). Trials are sorted by angular velocity. The trial starts at time = 0. White background indicates that the mouse is rotating, and a gray background indicates that the mouse is stationary. (Right) Tuning curve showing average rate (±SEM) as a function of angular velocity.
(F) Cartoon of linear non-linear Poisson (LNP) model. The model input variables rotation direction, and velocities are converted into a weight parameter. Then, they are converted by a non-linear exponential into a Poisson rate and matched to the observed data (see STAR Methods).
(G) Percentage of cells (average ± SEM) classified by the LNP model as rotation selective (CW and CCW cells) or tuned to AHV, as a function of significance threshold. Pie chart shows the percentage of classified cells using p ≤ 0.05.
(H) AHV tuning curve examples of three cells using the LNP model (average ± SEM).
(I) The average ± SEM tuning curve of all AHV cells classified by the LNP model.