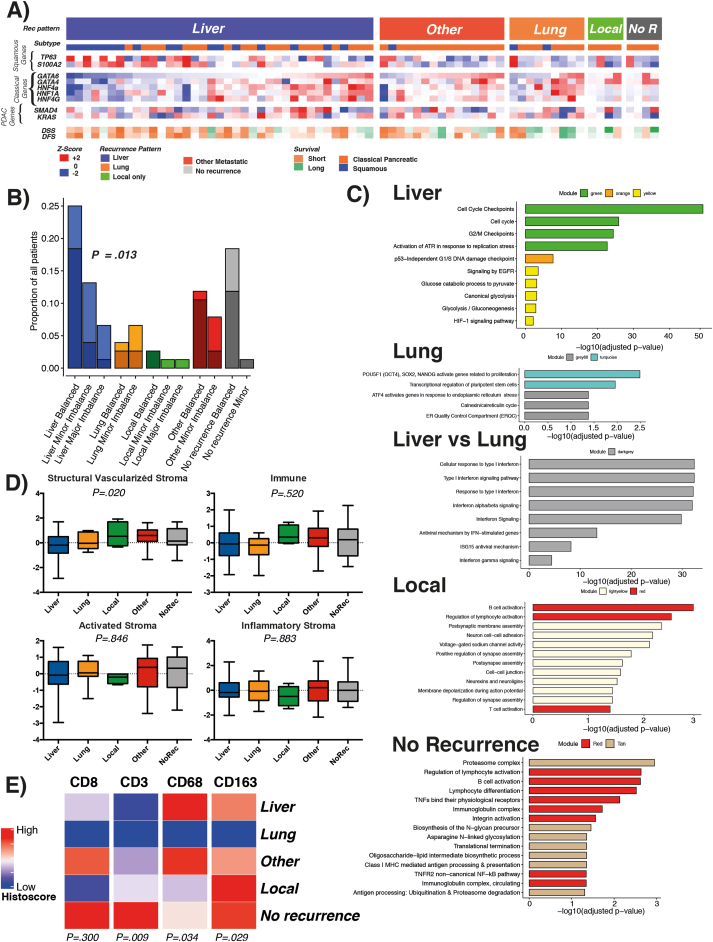
Figure 1.
(A) Heatmap of RNA sequenced cases with disease recurrence pattern. Heatmap demonstrates recurrence pattern, molecular subtype, relative expression of key genes of PC, classical and squamous subtype lineage, and outcome. (B) KRAS allelic imbalance and whole genome doubling in recurrence patterns. Scale is based on proportion of overall cohort. Dark shade represents no whole genome doubling and light shade whole genome doubling for each recurrence pattern stratified by KRAS allelic status. P value calculated for KRAS imbalance in liver versus all other recurrence patterns using chi-square test. (C) Relevant molecular pathways enriched in specific recurrence patterns categorized by gene module defined by Bailey et al.4 Size of bar proportional to statistical weight, horizontal scale numerical for P. (D) Stromal signatures as defined by Puleo et al7 for each recurrence pattern. P calculated as analysis of variance between groups. (E) Heatmap of relative immune cell infiltration histoscore in different recurrence patterns. Histoscore represents cumulative score for all tissue microarray cores per patient.