Abstract
Genome-wide association studies identified various loci associated with periodontal diseases, but assigning causal alleles remains difficult. Likewise, the generation of biological meaning underlying a statistical association has been challenging. Here, we characterized the genetic association at the gene ST8SIA1 that increases the risk for severe periodontitis in smokers. We used CRISPR/dCas9 activation and RNA-sequencing to identify genetic interaction partners of ST8SIA1 and to determine its function in the cell. We used reporter gene assays to identify regulatory elements at the associated single-nucleotide polymorphisms (SNPs) and to determine effect directions and allele-specific changes of enhancer activity. Antibody electrophoretic mobility shift assays proved allele-specific transcription factor binding at the putative causal SNPs. We found the reported periodontitis risk gene ABCA1 as the top upregulated gene following ST8SIA1 activation. Gene set enrichment analysis showed highest effects on integrin cell surface interactions (area under the curve [AUC] = 0.85; q = 4.9 × 10−6) and cell cycle regulation (AUC = 0.89; q = 1.6 × 10−5). We identified 2 associated repressor elements in the introns of ST8SIA1 that bind the transcriptional repressor BACH1. The putative causative variant rs2012722 decreased BACH1 binding by 40%. We also pinpointed ST8SIA1 as the target gene of the association. ST8SIA1 inhibits cell adhesion with extracellular matrix proteins, integrins, and cell cycle, as well as enhances apoptosis. Likewise, tobacco smoke reportedly results in an inhibition of cell adhesion and a decrease in integrin-positive cells and cell growth. We conclude that impaired ST8SIA1 repression, independently caused by reduced BACH1 binding at the effect T allele, as well as by tobacco smoke, contributes to higher ST8SIA1 levels, and in smokers who carry the effect T allele, both factors would be additive with damaging effects on the gingival barrier integrity. The activity of ST8SIA1 is also linked with the periodontitis risk gene ABCA1.
Keywords: periodontal diseases, alleles, tobacco smoke, reporter genes, extracellular matrix, transcription factors
Introduction
Genome-wide association studies (GWASs) identified numerous loci statistically associated with risk for complex human diseases and traits (Buniello et al. 2019). Complex diseases such as periodontitis are characterized by a complex etiology and genetic heterogeneity. The current challenge is to leverage the GWAS-implicated disease risk loci to biological meaning (Gallagher and Chen-Plotkin 2018). Several factors make it difficult to link the statistical associations to a functional understanding of the biology underlying disease risk. First, the associated regions often span thousands of DNA bases with the most significant disease-associated variant, called the lead single-nucleotide polymorphism (SNP) or sentinel variant, in strong linkage disequilibrium (LD) with many coinherited variants comprising an associated haplotype block (Gabriel et al. 2002). Consequently, the associations do not specify which of the linked variants at that locus is actually causing the association. This requires empirical validation to determine which variants are functional. Second, the molecular mechanism that the causal variant influences needs characterization to explain the role of the effect alleles in disease (Albert and Kruglyak 2015; Pai et al. 2015). Last, most of the GWAS-associated SNPs fall within nonprotein coding sequences. The different alleles of a SNP may cause variation in expression levels of messenger RNAs (mRNAs), if they are located in a gene regulatory noncoding chromatin element (Rockman and Kruglyak 2006). These genomic locations are termed expression quantitative trait loci (eQTL). It is considered that they influence the expression of either closely located (cis) or more remote (trans) genes (Bryois et al. 2014). Thus, an experimental approach to determine the actual target gene is necessary. Answering these problems connects the disease with a molecular mechanism and points to a regulatory genetic pathway. This improves our understanding of the disease etiology, and it might lead to new treatment options.
For a functional characterization of an association, complementary methods need to be integrated. CRISPR/dCas9 activation (CRISPRa) can be used to validate the physical interaction between a putative enhancer and a candidate gene promoter to determine the target gene of the association (Gilbert et al. 2014). Reporter gene assays allow direct testing of whether an associated variant locates to a regulatory sequence and whether an individual allele affects gene expression. An electrophoretic mobility shift assay (EMSA) allows interrogation of the binding affinities of a transcription factor in the presence of either the effect allele or the noneffect allele.
In a previous genome-wide gene × smoking interaction study, we identified a nominal genetic association at introns 1 and 2 of the gene ST8SIA1 (sialyltransferase ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1), a member of glycosyltransferase family 29, that increased the risk for periodontitis in smokers (Freitag-Wolf et al. 2019). For the disease-associated alleles, strong modifying effects on the expression of ST8SIA1 were reported by studies that investigated eQTLs (Zou et al. 2012; GTEx-Consortium 2013). The strong effects of these SNPs on the expression of ST8SIA1, which were much less for other genes, suggested ST8SIA1 as the target gene of the association. ST8SIA1 has a function in inhibition of cell adhesion with extracellular matrix (ECM) proteins, in S-phase arrest of the cell cycle and apoptosis (Mandal et al. 2014). Notably, ST8SIA1 showed strong upregulation after exposure to cigarette smoke extract in vitro (Freitag-Wolf et al. 2019). Tobacco smoke results in an inhibition of cell adhesion, a decrease in integrin-positive cells, and reduced cell growth (Semlali, Chakir, Goulet, et al. 2011; Semlali, Chakir, and Rouabhia 2011).
We hypothesized that this, in conjunction with the increased expression of ST8SIA1 after cigarette smoke exposure, made it an interesting candidate gene that might link the genetic susceptibility to periodontitis with the deleterious effects of tobacco smoke. The aim of the current study was to identify genes and gene networks that respond to increased ST8SIA1 expression and to characterize the association on the molecular level.
Materials and Methods
For clarity of the sequence of experiments, the workflow is illustrated in Figure 1.
Figure 1.
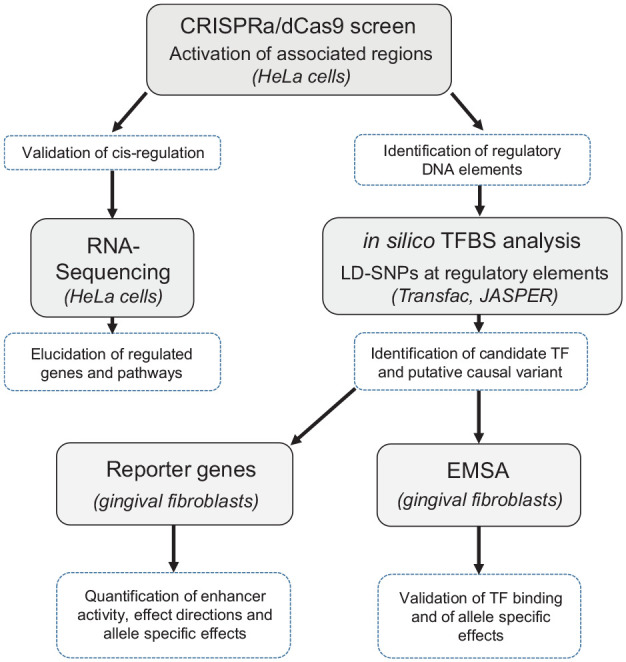
Workflow of the experiments. The shaded fields describe the methods. The material used is given in brackets. The dotted fields indicate putative findings that suggest the subsequent experiments.
Cell Culture, Quantitative Reverse Transcription Polymerase Chain Reaction, RNA-Sequencing, and Reporter Gene Assays
The protocols are described in the Appendix.
CRISPR-Mediated Gene Activation
We used CRISPRa to induce expression of ST8SIA1 for RNA-sequencing (RNA-seq). The advantage of CRISPRa compared to episomal expression plasmids is that it allows gene activation within the endogenous chromosomal context, including naturally occurring genetic variants and in physiological concentrations. CRISPRa also provides the possibility to test whether a genomic site serves as a cis-regulatory element for a target gene of interest. Therefore, in addition to the RNA-seq experiment, we used CRISPRa to validate if ST8SIA1 is the target gene of the gene × smoking association. A detailed description of the method is provided in the Appendix.
In Silico Analysis of Putative Causal Variants
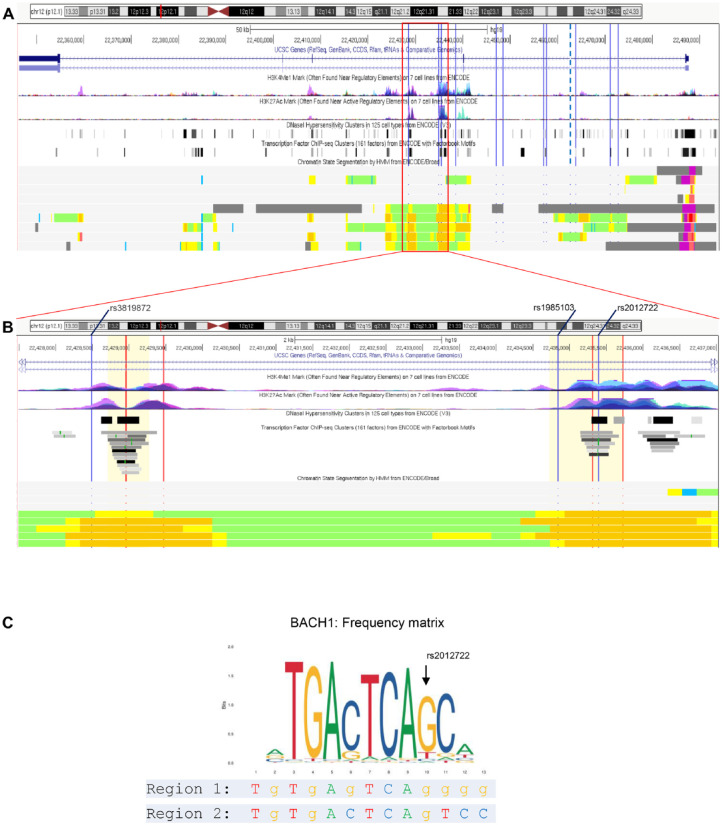
LD between the lead SNP rs2728821 and other common SNPs of this haplotype block was assessed using LDproxy Tool (Machiela and Chanock 2015) with population groups CEU (Utah Residents from North and West Europe) and GBR (British in England and Scotland). We assessed LD using r2 as a measure of correlation of alleles for 2 genetic variants (Appendix Table 1). We analyzed if these SNPs located to chromatin elements that correlate with regulatory functions of gene expression provided from ENCODE (ENCODE-Project-Consortium 2012) (Fig. 2). These elements were 1) open chromatin determined by DNAse I hypersensitivity (DHS), 2) H3K27Ac and H3K4Me1 histone modifications, 3) transcription factor binding sites (TFBSs) experimentally confirmed by chromatin immunoprecipitation sequencing (ChIP-seq), and 4) chromatin state segmentation, which combines epigenomic data into a sequence of functional chromatin states (Mammana and Chung 2015). To annotate eQTL effects of the associated SNPs, we used the software tool QTLizer (Munz et al. 2020). The eQTLs are listed in Appendix Table 2. To investigate whether these SNPs changed predicted TFBSs, we used the TF databases Transfac professional (geneXplain) (Wingender 2008) and the open-access database Jaspar (Sandelin et al. 2004).
Figure 2.
Genome-wide association study (GWAS)–nominated linkage disequilibrium (LD)–single-nucleotide polymorphisms (SNPs) and 2 putative regulatory regions at ST8SIA1. (A) The sentinel SNP (lead SNP) rs2728821 is marked with a bold dotted vertical line and the 11 LD-SNPs are marked with thin lines. From top: The first panel shows the chromosomal hg19 positions and the exon-intron structure of ST8SIA1. This gene is transcribed in reverse orientation with the promoter on the right. The second panel shows the ENCODE-derived H3K4me1 and H3K27ac methylation marks from 7 cell lines that are often associated with the higher activation of transcription and are defined as active enhancer marks. The third panel presents the ENCODE-derived DNase I hypersensitive sites from 125 cell types. These accessible chromatin zones are functionally related to transcriptional activity. The fourth panel presents the binding regions that were determined for 161 TFs by chromatin immunoprecipitation sequencing (ChIP-seq) experiments of ENCODE. This panel does not show the exact transcription factor binding site (TFBS) but indicates transcription factor (TF) binding was found at these chromatin regions. The bottom panel displays chromatin state segmentation for several human cell types (GM12878, H1-hESC, K562, HepG2, HUVEC, HMEC, HSMM, NHEK, NHLF) using a hidden Markov model (HMM) with ChIP-seq data for 9 TFs functionally related to transcriptional activity as input. The states are colored to highlight predicted functional elements (orange = strong enhancer, yellow = weak enhancer, green = weak transcribed, blue = insulator, red = active promoter, purple = poised promoter). (B) Closeup view of panel A. At 2 chromatin regions, several LD-SNPs locate within strong enhancers predicted by HMM chromatin state segmentation patterns. The chromosomal hg19 positions are given in the top panel. The 3 LD-SNPs are marked with blue lines. The regions covered by the reporter gene constructs are highlighted with lemon chiffon color. The positions of the single-guide RNAs (sgRNAs) are marked in red. (C) Frequency matrix (from Jaspar). The common G allele of SNP rs2012722 is predicted to be required for BACH1 binding, whereas the rare T allele strongly reduces binding affinity. Under the matrix, the DNA sequences of Region 1 (regulatory element tagged by rs3819872) and Region 2 (regulatory element tagged by rs2012722) at the predicted BACH1 binding motif are indicated.
Electrophoretic Mobility Shift Assay
Allele-specific oligonucleotide probes (Appendix Table 3) were designed to determine allele specificity of TF BTB and CNC homology 1 (BACH1) binding. The EMSA protocol is described in detail in the Appendix. The chemoluminescence of the blotted antibody was detected on an X-ray film for Western blot detection and quantified using the open-source image-processing program, ImageJ (Rueden et al. 2017).
Preparation and Induction of Cigarette Smoke Extract
The cigarette smoke extract (CSE) was prepared according to Freitag-Wolf et al. (2019). A detailed protocol is supplied in the Appendix.
Results
The Periodontitis-Associated Chromatin Elements Regulate ST8SIA1 Expression in cis
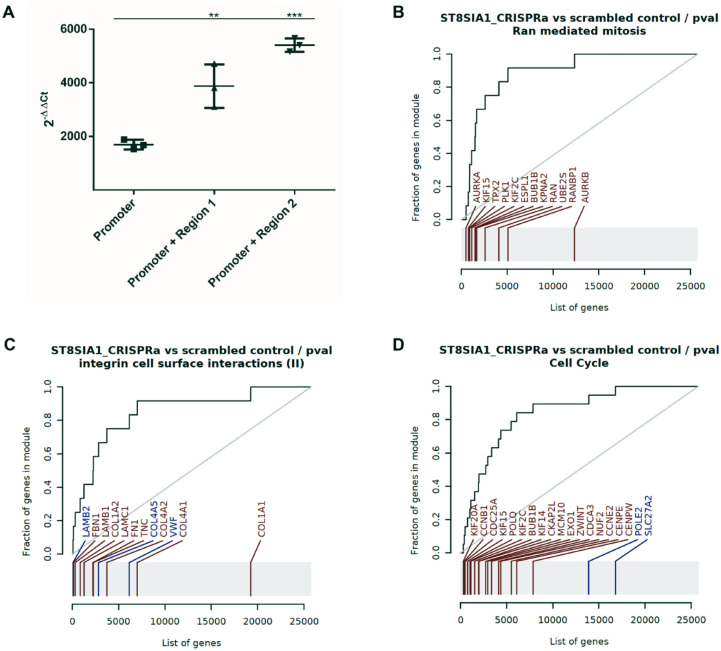
The reported eQTLs of the associated SNPs suggested cis-regulation of ST8SIA1 expression. However, assignment of regulatory effects of associated regions on specific genes based on eQLTs and linear proximity is error prone, because eQTLs are statistical observations with no direct molecular evidence (Zeng et al. 2017). Furthermore, enhancers often map large distances away from their actual targets and do not necessarily influence expression of the nearest gene. Induction of ST8SIA1 expression by positioning an activator protein to the locations of the associated SNPs by the CRISPRa system would prove their cis regulatory function. Cotransfection of HeLa cells with the CRISPRa system SAM (Synergistic Activation Mediators) and plasmids encoding single-guide RNAs (sgRNA) targeting the ST8SIA1 promoter increased the expression of ST8SIA1 1,694-fold (±180) compared to cotransfection with unspecific control sgRNAs. Cotransfection of promoter-targeting sgRNAs together with sgRNAs that targeted regulatory region at rs3819872 or rs2012722 further increased ST8SIA1 expression 3,877-fold (±808) and 5,403-fold (±253), respectively (Fig. 3A).
Figure 3.
Gene set enrichment analysis of CRISPRa induced ST8SIA1 expression in HeLa cells. (A) The periodontitis-associated DNA elements at Region 1 (regulatory element tagged by rs3819872) and Region 2 (regulatory element tagged by rs2012722) regulate ST8SIA1 expression. Data show 3 biological replicates for each experiment (**P = 0.0017, ***P = 0.0002). CRISPR-activated cells with guide RNAs (gRNAs) that targeted the promoter and regulatory region tagged by rs2012722 were used for RNA-sequencing (RNA-seq). (B–D) Evidence plots (receiver operator characteristic curves) for the top 3 gene sets. Each panel corresponds to 1 gene set. The gray rug plot underneath each curve corresponds to genes sorted by P value, with the genes belonging to the corresponding gene sets highlighted in red (upregulated genes) or blue (downregulated genes). Bright red or bright blue indicates that the genes are significantly regulated. The area under the curve (AUC) corresponds to the effect size of the enrichment, with 0.5 being no enrichment and 1.0 being maximal possible enrichment.
Overexpression of ST8SIA1 Upregulates ABCA1 and the Gene Sets “Mitosis” and “Integrin Cell Surface”
Exposure of cigarette smoke extract to gingival fibroblasts was shown to cause significant upregulation of ST8SIA1 expression (Freitag-Wolf et al. 2019). Here, we were interested in identifying ST8SIA1 responding genes and pathways. As the top upregulated gene (not counting ST8SIA1 itself), RNA-sequencing following endogenous activation of ST8SIA1 expression by CRISPRa found the suggestive periodontitis risk gene ATP binding cassette subfamily A member 1 (ABCA1) (Teumer et al. 2013) (Table). Gene set enrichment analysis using a second-generation algorithm, contrasting ST8SIA1 CRISPRa cells compared to scrambled sgRNA as controls, showed the highest effect sizes for the gene set “ran mediated mitosis” (LI.M15), with an area under the curve (AUC) = 0.89 (q = 1.6 × 10−5); “integrin cell surface interactions” (LI.M1.1), with an AUC = 0.85 (q = 4.9 × 10−6); and “cell cycle” (DC.M6.11), with AUC = 0.84 (q = 2.9 × 10−6) (Fig. 3B–D). In addition, GO enrichment analysis with hypergeometric test showed enrichment of the GO pathways “energy coupled proton transmembrane transport, against electrochemical gradient” (GO: 0015988) and “electron transport coupled proton transport” (GO:0015990), with q = 0.0016.
Table.
Top Up- and Downregulated Genes (P < 10−3) after CRISPRa of ST8SIA1 in HeLa Cells.
| Gene | Description | Fold Change (log2) | P Value | q Value |
|---|---|---|---|---|
| Upregulated genes | ||||
| ST8SIA1 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 9.5 | 1.7E-20 | 2.3E-16 |
| ABCA1 a | ATP binding cassette subfamily A member 1 | 1.6 | 4.2E-05 | 1.5E-02 |
| APCDD1L | APC downregulated 1 like | 1.5 | 4.9E-08 | 2.3E-04 |
| HLA-DMB | Major histocompatibility complex, class II, DM beta | 1.3 | 3.3E-05 | 1.4E-02 |
| CA11 | Carbonic anhydrase 11 | 1.3 | 1.6E-05 | 8.9E-03 |
| LPAR5 | Lysophosphatidic acid receptor 5 | 1.2 | 3.5E-07 | 5.7E-04 |
| HLA-DMA | Major histocompatibility complex, class II, DM alpha | 1.2 | 4.2E-07 | 5.7E-04 |
| ANGPTL2 | Angiopoietin-like 2 | 1.1 | 1.8E-04 | 3.9E-02 |
| Downregulated genes | ||||
| PKD1L1 | Polycystin 1 like 1, transient receptor potential channel interacting | −1.8 | 5.7E-05 | 1.9E-02 |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 | −0.7 | 5.9E-09 | 4.0E-05 |
| ENC1 | Ectodermal-neural cortex 1 | −0.5 | 5.2E-05 | 1.8E-02 |
| MXRA5 | Matrix remodeling associated 5 | −0.4 | 2.7E-05 | 1.3E-02 |
| ND5 | Mitochondrially encoded NADH/ubiquinone oxidoreductase core subunit 5 | −0.4 | 1.1E-05 | 6.8E-03 |
| AMH | Anti-Mullerian hormone | −0.4 | 3.9E-04 | 6.4E-02 |
| ND1 | Mitochondrially encoded NADH/ubiquinone oxidoreductase core subunit 1 | −0.4 | 1.1E-07 | 3.6E-04 |
| ZNF469 | Zinc finger protein 469 | −0.3 | 1.2E-04 | 3.1E-02 |
Periodontitis risk gene (Teumer et al. 2013), highlighted in bold letters.
Identification of Putative Causal Variants
SNP rs2728821 was reported as a lead SNP of the association at ST8SIA1 in the gene × smoking interaction analysis (Freitag-Wolf et al. 2019). rs2728821 is in strong LD (r2 > 0.8) with 11 additional coinherited variants. We tested whether the nucleotide variants of these 12 SNPs changed predicted TFBSs using Transfac professional and Jaspar. At rs2012722, Transfac professional predicted a TFBS for BACH1. This was confirmed by the motif collection of the database Jaspar. The rare T allele strongly reduced the binding affinity (frequency matrix [by Jaspar]: common allele = 13,050; rare allele = 1,545; i.e., 88.2% reduction; Fig. 2). Neither databases predicted different transcription factor (TF) binding affinities in the presence of any allele for the other SNPs in LD (Appendix Table 4). Of the 12 SNPs, rs2012722 alone located directly within a regulatory region (data from ENCODE).
BACH1 Has Reduced Binding Affinity at the Effect T Allele of rs2012722
To demonstrate protein binding at rs2012722, we performed an EMSA with allele-specific DNA probes. The band shift showed stronger protein binding in the background of the common G allele compared to the rare T allele (Fig. 4A, lanes 2 and 6). To give evidence of the specificity of BACH1 binding at rs2012722, we performed the EMSA with a BACH1-specific antibody. We observed a band super shift with the BACH1 antibody (lanes 3 and 7). In the background of the T allele, we observed 42% reduced BACH1 binding (Fig. 4B).
Figure 4.
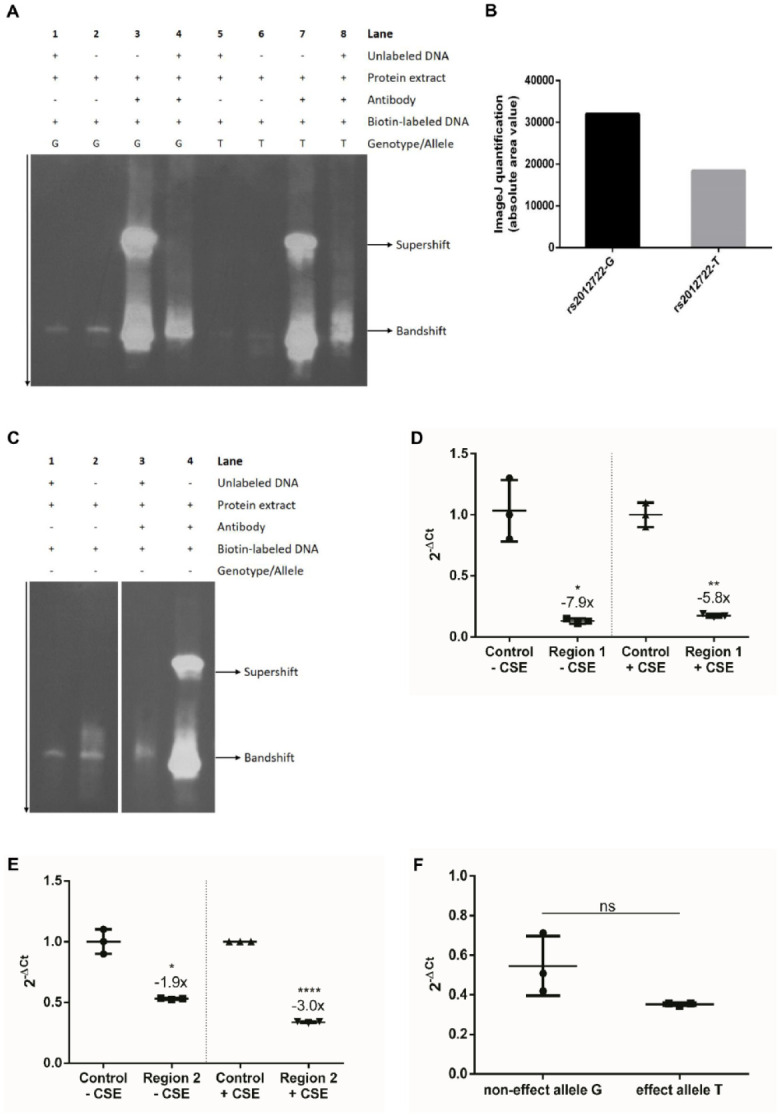
The disease-associated regulatory elements at ST8SIA1 show repressor activity and BACH1 binding. (A) Electrophoretic mobility shift assay (EMSA) indicated BACH1 binding at rs2012722 and revealed allele-specific effects on the binding affinity. Lanes 1 to 2 and 5 to 6 show EMSA without BACH1 antibody. The DNA probe that included the common G allele (lane 2) showed stronger protein binding than the DNA probe with the T allele (lane 6). Addition of unlabeled DNA showed the specificity of the band shift (lanes 1, 4, 5, and 8). The EMSA supershift was carried out by coincubation of an anti-Bach1 antibody with protein extract of gingival fibroblasts and DNA probes specific for the common G allele or the rare T allele of single-nucleotide polymorphism rs2012722. The effect T allele showed a weaker BACH1-AB supershift band compared to the noneffect G allele. (B) BACH1 binding affinity was 42% reduced in the background of the effect T allele compared to the noneffect G allele. (C) The band shift indicated BACH1 binding at rs3819872 (lanes 1–2, EMSA without BACH1 antibody). For the predicted BACH1 motif, the EMSA supershift was carried out by incubating an anti-Bach1 antibody with protein extract of gingival fibroblasts and specific DNA probes. The band supershift validated BACH1 binding at the predicted BACH1 transcription factor binding site (TFBS) (lane 4). (D, E) Reporter genes showed repressor activity of Region 1 (regulatory element tagged by rs3819872) and Region 2 (regulatory element tagged by rs2012722) in human immortalized fibroblasts 30 h after transfection independent of 6-h cigarette smoke extract (CSE) stimulation. (A) −CSE = −7.9-fold, P = 0.0228; +CSE = −5.8-fold, P = 0.0064. (B) −CSE = −1.9-fold, P = 0.0163; +CSE = −3.0-fold, P ≤ 0.0001. (F) Reporter gene plasmids with the repressor element (79 bp spanning rs2012722) showed reduced transcript levels compared to the control plasmid. The differences in reporter gene messenger RNA expression determined by quantitative reverse transcription polymerase chain reaction were not significant between the individual alleles (P > 0.05).
The Gene × Smoking Associated Haplotype Block Contains Multiple BACH1 Binding Sites
ENCODE data indicated the presence of an insulator separating the LD-SNPs downstream of rs2012722 (Fig. 2). Barrier insulators usually prevent silencing of euchromatin by the spread of neighboring heterochromatin (Gaszner and Felsenfeld 2006). The 7-kb upstream rs2012722 molecular features from ENCODE indicated a second 567–base pair (bp)–long regulatory region, tagged by disease-associated ST8SIA1 SNP rs3819872 (Fig. 2, Appendix Table 4). No insulator element was observed to separate both regions. At this putative regulatory region, no associated LD-SNP mapped to a predicted TFBS. It is considered that often the presence of more than 1 DNA binding site for a transcription factor in a regulatory region of a gene and the ability of the transcription factor to homodimerize contributes to regulation of the gene to a greater extent than the presence of a single DNA-binding site (Funnell and Crossley 2012). Therefore, it is conceivable that the presence of 2 (or more) DNA binding sites for a transcription factor results in synergistic effects of gene expression. We hypothesized that this regulatory region may also contain another BACH1 DNA binding motif. A search for this sequence motif across this DNA element identified the BACH1 binding motif at this region (chr12:22,428,944–56; hg19). To prove BACH1 binding at the DNA sequence of this predicted binding motif, we performed a supershift EMSA. This experiment validated BACH1 binding at this chromosomal position (Fig. 4C).
The Disease-Associated Regulatory Elements with BACH1 Binding Sites Are Transcriptional Repressors
Regulator elements can either activate or repress transcription. To measure the activity of the regulatory elements at the 2 BACH1 binding sites and to discriminate their effect directions, we employed reporter genes. Because we previously showed in vitro that ST8SIA1 responded with increased expression to exposure with cigarette smoke extract (Freitag-Wolf et al. 2019), we also tested whether the 2 regulatory elements responded to CSE. Twenty-four hours after cotransfection of gingival fibroblasts with the reporter gene plasmids for the 2 putative enhancers and of a reporter control plasmid, the transcript levels of the reporter genes for the regulatory regions tagged by rs3819872 and rs2012722 showed −7.9-fold change (P = 0.02) and −1.9-fold change reduction (P = 0.02), respectively (Fig. 4D–E). CSE stimulation did not significantly change repressor activity compared to the no-CSE controls.
Discussion
Using CRISPRa, we showed that the identified periodontitis-associated repressor elements regulate ST8SIA1 expression, implying ST8SIA1 as target gene of the association. ST8SIA1 is a membrane protein that is responsible for the production of gangliosides (GDs), specifically of GD3, which has a particular role in cell adhesion (Sasaki et al. 1994). We showed that ST8SIA1 activation by CRISPRa induced several gene sets, and the highest effect sizes were “mitosis,” “integrin cell surface interactions,” and “cell cycle.” This is concordant with another study that showed ST8SIA1-transfected pancreatic cancer cells exhibited inhibition of cell adhesion with ECM proteins and showed arrest in the S-phase of the cell cycle as well as enhanced apoptosis (Mandal et al. 2014). These findings imply a function of ST8SIA1 in regulation of integrin-mediated cell adhesion in formation and remodeling of the ECM.
We identified a TFBS for the transcription factor BACH1 at the associated ST8SIA1 SNP rs2012722 and confirmed BACH1 binding, implying a functional role of BACH1 signaling for the association with periodontitis and this SNP as the causal variant. BACH1 is widely expressed in most tissues and functions primarily as a transcriptional suppressor. It has known roles in the regulation of cell cycle and mitosis, and it suppresses angiogenesis by regulating target genes of the Wnt/β-catenin signaling pathway (Zhang et al. 2018). We showed an allele-specific effect on the TF binding affinity, using a BACH1 antibody in a supershift EMSA. However, unlike the supershift EMSA with the BACH1 antibody, the reporter assays did not show allele-specific transcript changes. This could indicate that the effect of the effect T allele of rs2012722 was below the detection limit of the reporter gene. However, while the DNA probe of the EMSA was 43 bp in length, the enhancer sequences of the reporter genes comprised 79 bp. It is possible that additional TFs present in the protein extract of the gingival fibroblasts bound to the reporter genes and stabilized BACH1 binding, thus compensating the effect of the causative T allele in this in vitro system. In summary, the supershift EMSA and the short probe were better able to identify the allele-specific effect of the putative causative variant of rs2012722, but the sensitivity of the reporter gene system was adequate to identify directional effect of the enhancers.
Dysregulation of ST8SIA1 could be harmful on gingival barrier integrity and wound healing (Mandal et al. 2014). The effect T allele reduced binding of the repressor BACH1, and CSE exposure significantly upregulated ST8SIA1 expression. However, CSE showed no significant effect on the activity of the reporter gene that included the repressor element at rs2012722. Thus, increased ST8SIA1 expression due to smoking and the risk allele would be independent effects. This implies that the effects of the risk T allele and smoking were additive in their contribution to unlock ST8SIA1 repression. Similar to ST8SIA1 function, tobacco smoke causes inhibition of cell adhesion, decreased numbers of β1-integrin-positive cells, and reduced cell growth (Semlali, Chakir, Goulet, et al. 2011). Smoke-exposed fibroblasts were not able to contract collagen gel matrix and migrate following CSE exposure, which may negatively affect periodontal wound healing (Semlali, Chakir, Rouabhia 2011). These effects of tobacco smoking would add to similar effects of ST8SIA1. Taken together, it can be speculated that the effects of tobacco smoking in carriers of the effect T allele are additive. In contrast, nonsmokers may be able to compensate the effect of the risk allele, because the nonsmoking patient group showed no association with increased disease susceptibility and the risk haplotype.
Interestingly, CRISPRa upregulation of ST8SIA1 showed that the gene ABCA1 was the top upregulated gene. Studies with ABCA1 knockout mice demonstrated anti-inflammatory roles for this transporter (Zhu et al. 2008; Tang et al. 2009). A GWAS on periodontitis reported ABCA1 as a suggestive risk gene of periodontitis, with rs4149263-A associated withP = 7 × 10−6, odds ratio = 0.8 (95% confidence interval, 0.03–0.08) (Teumer et al. 2013). We conclude that both genes are located in the same regulatory cascade, implying a relevant role of this molecular network in the etiology of periodontitis. This network could contribute to impaired barrier integrity at minor signs of inflammation.
Limitations of this study were that we possibly missed TFBSs. Approaches to model the binding specificity and position weight matrix (PWM) of a TF have limitations, because the specificity of protein-DNA binding also depends on the 3-dimensional structure of DNA and TF protein macromolecules and not only on the DNA sequence (Rohs et al. 2010), and TF binding motifs are not strictly conserved. This results in different motif sequences and limited predictive accuracy of PWMs (Weirauch et al. 2013). In addition, we analyzed only SNPs in strong linkage (r2 > 0.8). We did this because the r2 coefficient of correlation takes account of allele frequency. Strong LD, indicated by D′ but not by r2, would include alleles that are inherited with the particular lead SNP but are not carried by the majority of cases because they are rare (Slatkin 2008). Such alleles would not be suggestive as causative variants because they would not explain the association for the majority of cases. Another limitation was that CRISPRa and RNA-seq was performed in HeLa cells instead of gingival cells. This was done because gingival fibroblasts and epithelial cells showed poor survival after transfection with the CRISPRa plasmids, probably because of DNA toxicity, whereas HeLa cells showed high survival after transfection of the CRISPRa system. We hypothesized that recruitment of a strong transcriptional activator to an enhancer would be sufficient to drive target gene expression, even if that enhancer was not currently active in the assayed cells as recently demonstrated (Simeonov et al. 2017). HeLa cells are of different developmental origin than gingival cells, and it is possible that their transcriptional response to ST8SIA1 upregulation differs from gingival cells. But the observed effects were similar to previous overexpression of ST8SIA1 in pancreatic cancer cells (Mandal et al. 2014). Thus, we consider HeLa cells as an appropriate cell model for our CRISPRa experiments.
In summary, we showed that the periodontitis risk genes ST8SIA1 and ABCA1 locate to the same genetic pathway. We also identified and characterized the putative causal variant of the gene × smoking interaction at ST8SIA1 and pinpointed this glycosyltransferase gene as the target gene of the association with severe periodontitis.
Author Contributions
A. Chopra, A.S. Schaefer, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; R. Mueller, J. Weiner 3rd, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; J. Rosowski, H. Dommisch, E. Grohmann, contributed to data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211017510 for BACH1 Binding Links the Genetic Risk for Severe Periodontitis with ST8SIA1 by A. Chopra, R. Mueller, J. Weiner 3rd, J. Rosowski, H. Dommisch, E. Grohmann and A.S. Schaefer in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by a PhD fellowship of Beuth Hochschule für Technik Berlin to A. Chopra and by a grant of the German Research foundation (DFG) to A.S. Schaefer (SCH 1582/5-1).
RNA-Seq Data: Reads, raw counts, and results of differential expression analysis have been submitted to the Short Read Archive via Gene Expression Omnibus (GEO accession GSE160672).
ORCID iDs: A. Chopra  https://orcid.org/0000-0001-7706-3577
https://orcid.org/0000-0001-7706-3577
H. Dommisch  https://orcid.org/0000-0003-1043-2651
https://orcid.org/0000-0003-1043-2651
References
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 16(4):197–212. [DOI] [PubMed] [Google Scholar]
- Bryois J, Buil A, Evans DM, Kemp JP, Montgomery SB, Conrad DF, Ho KM, Ring S, Hurles M, Deloukas P, et al. 2014. Cis and trans effects of human genomic variants on gene expression. PLoS Genet. 10(7):e1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. 2019. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47(D1):D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE-Project-Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag-Wolf S, Munz M, Wiehe R, Junge O, Graetz C, Jockel-Schneider Y, Staufenbiel I, Bruckmann C, Lieb W, Franke A, et al. 2019. Smoking modifies the genetic risk for early-onset periodontitis. J Dent Res. 98(12):1332–1339. [DOI] [PubMed] [Google Scholar]
- Funnell APW, Crossley M. 2012. Homo- and heterodimerization in transcriptional regulation. In: Matthews JM, editor. Protein dimerization and oligomerization in biology. New York, NY: Springer New York. p. 105–121. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. 2002. The structure of haplotype blocks in the human genome. Science. 296(5576):2225–2229. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Chen-Plotkin AS. 2018. The post-GWAS era: from association to function. Am J Hum Genet. 102(5):717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 7(9):703–713. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 159(3):647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx-Consortium. 2013. The genotype-tissue expression (GTEx) project. Nat Genet. 45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. 2015. Ldlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammana A, Chung HR. 2015. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol. 16(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal C, Sarkar S, Chatterjee U, Schwartz-Albiez R, Mandal C. 2014. Disialoganglioside GD3-synthase over expression inhibits survival and angiogenesis of pancreatic cancer cells through cell cycle arrest at S-phase and disruption of integrin-β1-mediated anchorage. Int J Biochem Cell Biol. 53:162–173. [DOI] [PubMed] [Google Scholar]
- Munz M, Wohlers I, Simon E, Reinberger T, Busch H, Schaefer AS, Erdmann J. 2020. Qtlizer: comprehensive QTL annotation of GWAS results. Sci Rep. 10(1):20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Pritchard JK, Gilad Y. 2015. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 11(1):e1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. 2006. Genetics of global gene expression. Nat Rev Genet. 7(11):862–872. [DOI] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. 2010. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 18(1):529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. 2004. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32(Database issue):D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Kurata K, Kojima N, Kurosawa N, Ohta S, Hanai N, Tsuji S, Nishi T. 1994. Expression cloning of a GM3-specific alpha-2,8-sialyltransferase (GD3 synthase). J Biol Chem. 269(22):15950–15956. [PubMed] [Google Scholar]
- Semlali A, Chakir J, Goulet JP, Chmielewski W, Rouabhia M. 2011. Whole cigarette smoke promotes human gingival epithelial cell apoptosis and inhibits cell repair processes. J Periodontal Res. 46(5):533–541. [DOI] [PubMed] [Google Scholar]
- Semlali A, Chakir J, Rouabhia M. 2011. Effects of whole cigarette smoke on human gingival fibroblast adhesion, growth, and migration. J Toxicol Environ Health A. 74(13):848–862. [DOI] [PubMed] [Google Scholar]
- Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, et al. 2017. Discovery of stimulation-responsive immune enhancers with crispr activation. Nature. 549(7670):111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 2008. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 9(6):477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Liu Y, Kessler PS, Vaughan AM, Oram JF. 2009. The macrophage cholesterol exporter abca1 functions as an anti-inflammatory receptor.J Biol Chem. 284(47):32336–32343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teumer A, Holtfreter B, Volker U, Petersmann A, Nauck M, Biffar R, Volzke H, Kroemer HK, Meisel P, Homuth G, et al. 2013. Genome-wide association study of chronic periodontitis in a general German population. J Clin Periodontol. 40(11):977–985. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, et al. 2013. Evaluation of methods for modeling transcription factor sequence specificity. Nat Biotechnol. 31(2):126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. 2008. The transfac project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 9(4):326–332. [DOI] [PubMed] [Google Scholar]
- Zeng B, Lloyd-Jones LR, Holloway A, Marigorta UM, Metspalu A, Montgomery GW, Esko T, Brigham KL, Quyyumi AA, Idaghdour Y, et al. 2017. Constraints on eQTL fine mapping in the presence of multisite local regulation of gene expression. G3 (Bethesda). 7(8):2533–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo J, Wei X, Niu C, Jia M, Li Q, Meng D. 2018. Bach1: function, regulation, and involvement in disease. Oxid Med Cell Longev. 2018:1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Lee JY, Timmins JM, Brown JM, Boudyguina E, Mulya A, Gebre AK, Willingham MC, Hiltbold EM, Mishra N, et al. 2008. Increased cellular free cholesterol in macrophage-specific Abca1 knock-out mice enhances pro-inflammatory response of macrophages. J Biol Chem. 283(34):22930–22941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al. 2012. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 8(6):e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211017510 for BACH1 Binding Links the Genetic Risk for Severe Periodontitis with ST8SIA1 by A. Chopra, R. Mueller, J. Weiner 3rd, J. Rosowski, H. Dommisch, E. Grohmann and A.S. Schaefer in Journal of Dental Research